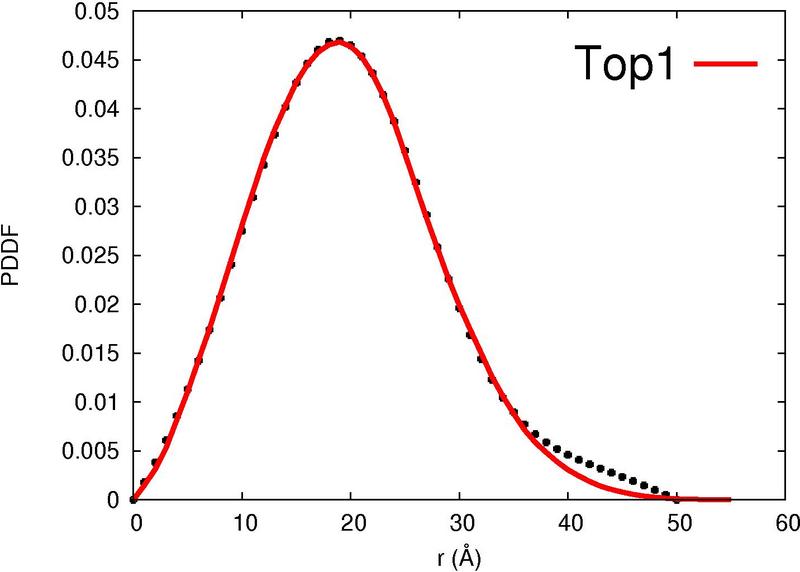
 | 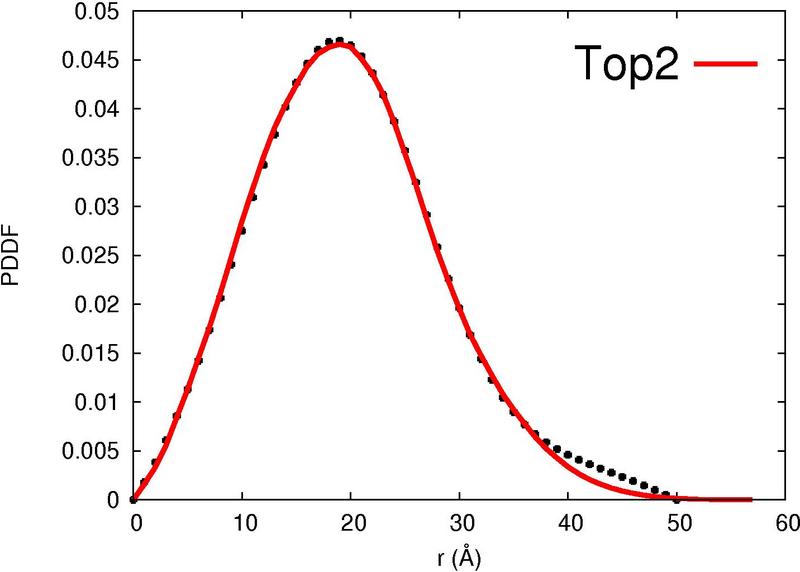 | 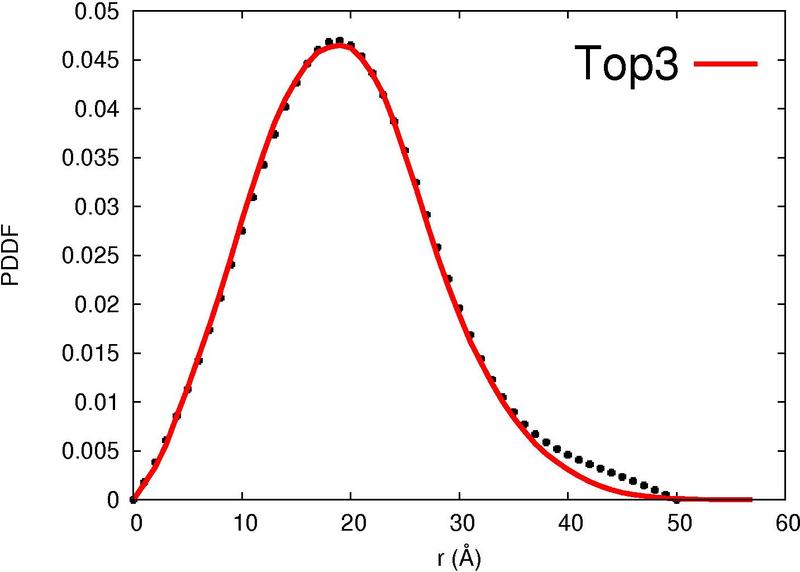 | 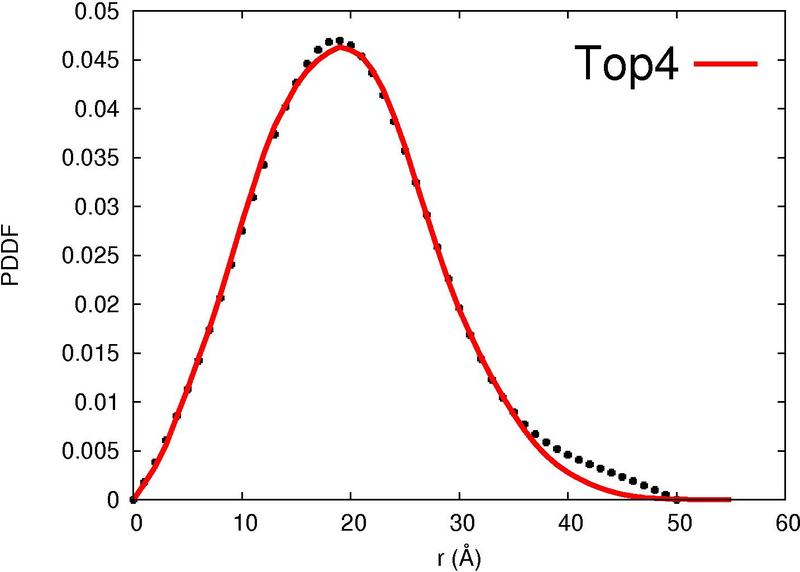 |  |
| Download profile 1 | Download profile 2 | Download profile 3 | Download profile 4 | Download profile 5 |
 |  |  |  |  |
| Download profile 6 | Download profile 7 | Download profile 8 | Download profile 9 | Download profile 10 |
| Seq: | KVFGRCELAAAMKRHGLDNYRGYSLGNWVCAAKFESNFNTQATNRNTDGSTDYGILQINS |
| SS : | CCCCHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHCCCCCCEECCCCCCCCCCCEEECC |
| Pos: | 123456789012345678901234567890123456789012345678901234567890 |
| Seq: | RWWCNDGRTPGSRNLCNIPCSALLSSDITASVNCAKKIVSDGNGMNAWVAWRNRCKGTDV |
| SS : | HHHCCCCCCCCCCCCCCCCHHHHCCCCHHHHHHHHHHHHHCCCCCCHHHHHHHHCCCCCC |
| Pos: | 123456789012345678901234567890123456789012345678901234567890 |
| Seq: | QAWIRGCRL |
| SS : | HHHHHCCCC |
| Pos: | 123456789 |
 |  |  |  |  |
| Download profile 1 | Download profile 2 | Download profile 3 | Download profile 4 | Download profile 5 |
 |  |  |  |  |
| Download profile 6 | Download profile 7 | Download profile 8 | Download profile 9 | Download profile 10 |
| Rank | Template | Align_length | Coverage | Seq_id | Z-score threading | SAXS-score | SAXSTER score | Confidence | Target-template alignments | 3-D models from threading alignments | alpha-Carbon trace from I-TASSER | Full-atomic models by MODELLER |
| 1 | 1e8lA | 129 | 1.000 | 1.000 | 29.129 | 3.196 | 21.641 | High | alignment_1 | threading_1 | CAtrace_1 | model_1 |
| 2 | 1di3A | 129 | 1.000 | 0.605 | 28.907 | 3.16 | 21.47 | High | alignment_2 | threading_2 | CAtrace_2 | model_2 |
| 3 | 2z2fA | 128 | 0.992 | 0.547 | 28.344 | 3.067 | 21.034 | High | alignment_3 | threading_3 | CAtrace_3 | model_3 |
| 4 | 2gv0A | 129 | 1.000 | 0.690 | 28.28 | 2.992 | 20.949 | High | alignment_4 | threading_4 | CAtrace_4 | model_4 |
| 5 | 2eqlA | 128 | 0.992 | 0.492 | 27.992 | 3.177 | 20.854 | High | alignment_5 | threading_5 | CAtrace_5 | model_5 |
| 6 | 2goiA | 127 | 0.984 | 0.504 | 28.123 | 2.957 | 20.823 | High | alignment_6 | threading_6 | CAtrace_6 | model_6 |
| 7 | 1jugA | 125 | 0.969 | 0.536 | 27.902 | 3.125 | 20.764 | High | alignment_7 | threading_7 | CAtrace_7 | model_7 |
| 8 | 1iizA | 119 | 0.922 | 0.412 | 25.15 | 3.439 | 19.06 | High | alignment_8 | threading_8 | CAtrace_8 | model_8 |
| 9 | 1fkqA | 122 | 0.946 | 0.418 | 24.497 | 3.846 | 18.839 | High | alignment_9 | threading_9 | CAtrace_9 | model_9 |
| 10 | 1hfxA | 123 | 0.953 | 0.325 | 24.287 | 3.299 | 18.394 | High | alignment_10 | threading_10 | CAtrace_10 | model_10 |
| Column 1 : | The rank of the template. The template with the highest SAXSTER score is ranked 1. |
| Column 2 : | Name of the template PDB from which the particular model was generated. |
| Column 3 : | The length of the threading alignment between the query sequence and the template. |
| Column 4 : | The coverage of the alignment is calculated by dividing the length of the alignment (previous column) by the total length of the query sequence. |
| Column 5 : | The sequence identity in the aligned region between the query and the template sequences |
| Column 6 : | Z-score, or statistical significance of the threading alignment generated by MUSTER. |
| Column 7 : | SAXS-score indicates the correlation between the experimental curve and the theoretical SAXS profile of the template. |
| Column 8 : | SAXSTER score is the combined score between Z-score of the threading alignment and the SAXS-score. Templates are ranked according to the SAXSTER score. |
| Column 9 : | The confidence of the SAXSTER prediction is based on the Z-score of the alignment and the SAXS profile fitting from SAXSTER benchmarking results. The confidence is considered High if the SAXSTER score is greater than 6.3 or Low otherwise. |
| Column 10 : | The threading alignment between the query sequence and the template. |
| Column 11 : | The models generated from MUSTER threading and re-ranked by SAXSTER. |
| Column 12 : | The alpha-Carbon trace generated by I-TASSER from the threading alignment. This model is used to derive the SAXS profile from a coarse-grained simulation. |
| Column 13 : | Full-atomic models obtained by MODELLER from the alpha-Carbon trace generated by I-TASSER. |