

You can:
| Name | Neuromedin-U receptor 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | NMUR1 |
| Synonym | NMU1 receptor NmU-R1 GPR66 G-protein coupled receptor FM-3 G-protein coupled receptor 66 [ Show all ] |
| Disease | N/A |
| Length | 426 |
| Amino acid sequence | MTPLCLNCSVLPGDLYPGGARNPMACNGSAARGHFDPEDLNLTDEALRLKYLGPQQTELFMPICATYLLIFVVGAVGNGLTCLVILRHKAMRTPTNYYLFSLAVSDLLVLLVGLPLELYEMWHNYPFLLGVGGCYFRTLLFEMVCLASVLNVTALSVERYVAVVHPLQARSMVTRAHVRRVLGAVWGLAMLCSLPNTSLHGIRQLHVPCRGPVPDSAVCMLVRPRALYNMVVQTTALLFFCLPMAIMSVLYLLIGLRLRRERLLLMQEAKGRGSAAARSRYTCRLQQHDRGRRQVTKMLFVLVVVFGICWAPFHADRVMWSVVSQWTDGLHLAFQHVHVISGIFFYLGSAANPVLYSLMSSRFRETFQEALCLGACCHRLRPRHSSHSLSRMTTGSTLCDVGSLGSWVHPLAGNDGPEAQQETDPS |
| UniProt | Q9HB89 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9HB89 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9HB89. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL1075178 |
| IUPHAR | 298 |
| DrugBank | N/A |
| Name | CHEMBL3315348 |
|---|---|
| Molecular formula | C51H67N15O8 |
| IUPAC name | (2S)-2-[[(2S)-5-(diaminomethylideneamino)-2-[[(2S)-1-[(2S)-5-(diaminomethylideneamino)-2-[[(2S)-2-[[(2S)-3-naphthalen-2-yl-2-(3-pyridin-3-ylpropanoylamino)propanoyl]amino]-3-phenylpropanoyl]amino]pentanoyl]pyrrolidine-2-carbonyl]amino]pentanoyl]amino]butanediamide |
| Molecular weight | 1018.19 |
| Hydrogen bond acceptor | 11 |
| Hydrogen bond donor | 11 |
| XlogP | -0.7 |
| Synonyms | GTPL7926 BDBM50049424 compound 8d [PMID: 24999562] D0M9DD |
| Inchi Key | HPJGEESDHAUUQR-SKGSPYGFSA-N |
| Inchi ID | InChI=1S/C51H67N15O8/c52-42(67)29-38(44(53)69)64-45(70)36(15-7-23-59-50(54)55)62-48(73)41-17-9-25-66(41)49(74)37(16-8-24-60-51(56)57)63-47(72)40(27-31-10-2-1-3-11-31)65-46(71)39(61-43(68)21-19-32-12-6-22-58-30-32)28-33-18-20-34-13-4-5-14-35(34)26-33/h1-6,10-14,18,20,22,26,30,36-41H,7-9,15-17,19,21,23-25,27-29H2,(H2,52,67)(H2,53,69)(H,61,68)(H,62,73)(H,63,72)(H,64,70)(H,65,71)(H4,54,55,59)(H4,56,57,60)/t36-,37-,38-,39-,40-,41-/m0/s1 |
| PubChem CID | 91623352 |
| ChEMBL | CHEMBL3315348 |
| IUPHAR | 7926 |
| BindingDB | 50049424 |
| DrugBank | N/A |
Structure | 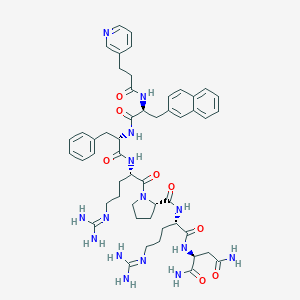 |
| Lipinski's druglikeness | This ligand has more than 5 hydrogen bond donor. This ligand has more than 10 hydrogen bond acceptor. This ligand is heavier than 500 daltons. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 5.1 nM | PMID24999562 | ChEMBL |
| Activity | 63.0 % | PMID24999562 | ChEMBL |
| Activity | 84.0 % | PMID24999562 | ChEMBL |
| EC50 | 1.0 - 10.0 nM | PMID24999562 | IUPHAR |
| EC50 | 2.2 nM | PMID24999562 | BindingDB,ChEMBL |
| Intrinsic activity | 79.0 % | PMID24999562 | ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218