

You can:
| Name | D(2) dopamine receptor |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Drd2 |
| Synonym | D2 receptor D2(415) and D2(444) D2A and D2B D2R Dopamine D2 receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 444 |
| Amino acid sequence | MDPLNLSWYDDDLERQNWSRPFNGSEGKADRPHYNYYAMLLTLLIFIIVFGNVLVCMAVSREKALQTTTNYLIVSLAVADLLVATLVMPWVVYLEVVGEWKFSRIHCDIFVTLDVMMCTASILNLCAISIDRYTAVAMPMLYNTRYSSKRRVTVMIAIVWVLSFTISCPLLFGLNNTDQNECIIANPAFVVYSSIVSFYVPFIVTLLVYIKIYIVLRKRRKRVNTKRSSRAFRANLKTPLKGNCTHPEDMKLCTVIMKSNGSFPVNRRRMDAARRAQELEMEMLSSTSPPERTRYSPIPPSHHQLTLPDPSHHGLHSNPDSPAKPEKNGHAKIVNPRIAKFFEIQTMPNGKTRTSLKTMSRRKLSQQKEKKATQMLAIVLGVFIICWLPFFITHILNIHCDCNIPPVLYSAFTWLGYVNSAVNPIIYTTFNIEFRKAFMKILHC |
| UniProt | P61169 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL339 |
| IUPHAR | 215 |
| DrugBank | N/A |
| Name | DIHYDREXIDINE |
|---|---|
| Molecular formula | C17H17NO2 |
| IUPAC name | (6aR,12bS)-5,6,6a,7,8,12b-hexahydrobenzo[a]phenanthridine-10,11-diol |
| Molecular weight | 267.328 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 3 |
| XlogP | 2.5 |
| Synonyms | (6aR,12bS)-5,6,6a,7,8,12b-hexahydrobenzo[a]phenanthridine-10,11-diol Benzo(a)phenanthridine-10,11-diol, 5,6,6a,7,8,12b-hexahydro-, (6ar-trans)- Q3PJ4B4D0X 5,6,6a,7,8,12b-Hexahydro-benzo[a]phenanthridine-10,11-diol CCG-204474 [ Show all ] |
| Inchi Key | BGOQGUHWXBGXJW-RHSMWYFYSA-N |
| Inchi ID | InChI=1S/C17H17NO2/c19-15-7-10-5-6-14-17(13(10)8-16(15)20)12-4-2-1-3-11(12)9-18-14/h1-4,7-8,14,17-20H,5-6,9H2/t14-,17-/m1/s1 |
| PubChem CID | 6603820 |
| ChEMBL | CHEMBL25856 |
| IUPHAR | N/A |
| BindingDB | 50010686 |
| DrugBank | N/A |
Structure | 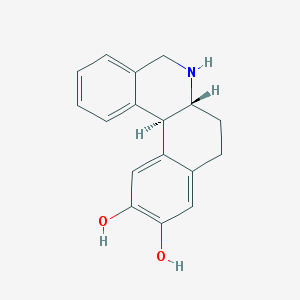 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | 70.0 nM | PMID17154515 | BindingDB,ChEMBL |
| IC50 | 87.7 nM | PMID7914538 | ChEMBL |
| IC50 | 107.0 nM | PMID7830274 | BindingDB,ChEMBL |
| IC50 | 120.0 nM | PMID1971308 | BindingDB,ChEMBL |
| IC50 | 132.0 nM | PMID7914538 | BindingDB,ChEMBL |
| IC50 | 1250.0 nM | PMID7914538 | BindingDB,ChEMBL |
| Intrinsic activity | 70.0 nM | PMID17154515 | ChEMBL |
| K 0.5 | 58.1 nM | PMID10072690 | ChEMBL |
| K 0.5 | 1490.0 nM | PMID10072690 | ChEMBL |
| K0.5 | 29.0 nM | PMID7608904 | ChEMBL |
| K0.5 | 43.2 nM | PMID8558526 | ChEMBL |
| K0.5 | 43.8 nM | PMID8568818, PMID9216832 | ChEMBL |
| K0.5 | 58.1 nM | PMID7636869 | ChEMBL |
| Kd | 2300.0 nM | PMID10956209 | BindingDB,ChEMBL |
| Kd | 2500.0 nM | PMID10956209 | BindingDB,ChEMBL |
| Kd | 3500.0 nM | PMID10956209 | BindingDB,ChEMBL |
| Kd | 13000.0 nM | PMID10956209 | BindingDB,ChEMBL |
| Ki | 1400.0 nM | PMID9171869 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218