| Synonyms | Risperidone solution, 1.0 mg/mL in methanol, ampule of 1 mL, certified reference material
Risperidone, Pharmaceutical Secondary Standard; Certified Reference Material
Rispolin
SPBio_003078
Tox21_110253
NSC-759895
Q-4040
R-64-766
Risperdal
Risperidona [Spanish]
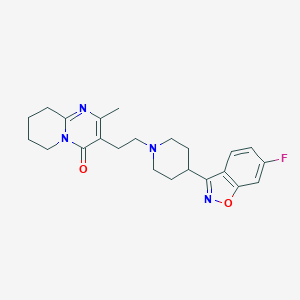
3-[2-[4(6-fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl]ethyl]-6,7,8,9-tetrahydro-2-methyl4H-pyrido[1,2-a]pyrimidin-4-one
3-[2-[4-(6-Fluoro-1,2-benzisoxazol-3-yl)piperi-dino]ethyl]-6,7,8,9-tetrahydro-2-methyl-4H-pyrido[1,2-a]-pyrimidin-4-one
3-{2-[4-(6-fluoro-1,2-benzisoxazol-3-yl)piperidin-1-yl]ethyl}-2-methyl-6,7,8,9-tetrahydro-4H-pyrido[1,2-a]pyrimidin-4-one
GTPL96
3-{2-[4-(6-Fluoro-benzo[d]isoxazol-3-yl)-piperidin-1-yl]-ethyl}-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one
HMS3657G13
4767-EP2280010A2
KS-00000760
4H-Pyrido(1,2-a)pyrimidin-4-one, 3-(2-(4-(6-fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl)ethyl)-6 ,7,8,9-tetrahydro-2-methyl-
MLS000759429
NCGC00015883-04
AB00514010_14
API0004081
2-(2-(4-(benzo[d]isoxazol-3-yl)piperidin-1-yl)ethyl)-7,8,9,9a-tetrahydro-1H-pyrido[1,2-a]pyrimidin-4(6H)-one
BRD-K53857191-001-10-2
Risperidone (JP17/USAN/INN)
Risperidone(R 64 766)
Risperidone-ISM
SAM001246595
SR-01000075399-2
UNII-L6UH7ZF8HC
Pharmakon1600-01506038
R 64 766, Risperdal, Risperidone
R64,766
Risperdal M-Tab
3-[2-[4-(6-fluoro- 1,2-benzisoxazol-3-yl)-1-piperidinyl]ethyl]-6,7,8,9-tetrahydro-2-methyl4H-pyrido[1,2-a]pyrimidin-4-one
CPD000466323
3-[2-[4-(6-fluoro-1,2-benzoxazol-3-yl)-1-piperidyl]ethyl]-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one
DSSTox_GSID_45193
3-{2-[4-(6-fluoro-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl}-2-methyl-6,7,8,9-tetrahydro-4H-pyrido[1,2-a]pyrimidin-4-one
HMS2089C22
4767-EP2272537A2
HY-11018
4767-EP2301936A1
L6UH7ZF8HC
4H-Pyrido[1,2-a]pyrimidin-4-one, 3-[2-[4-(6-fluoro-1,2-benz-isoxazol-3-yl)-1-piperidinyl]ethyl]-6,7,8,9-tetrahydro-2-methyl-
MolPort-002-885-858
AB00514010
NCGC00015883-07
ACT04270
BDBM50001885
3-(2-(4-(6-Fluorobenzo[d]isoxazol-3-yl)piperidin-1-yl)ethyl)-2-methyl-6,7,8,9-tetrahydro-4H-pyrido[1,2-a]pyrimidin-4-one
C23H27FN4O2
Risperidone 1.0 mg/ml in Methanol
Risperidone, British Pharmacopoeia (BP) Reference Standard
Risperin
Sequinan
SW197348-4
NCGC00179257-01
ZINC538312
Psychodal
R-118
Relday
Risperidal M-Tab
CCG-100930
3-[2-[4-(6-fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl]ethyl]-6,7,8,9-tetrahydro-2-methyl-4H-pyrido[1,2-a]pyrimidin-4-one
D00426
3-{2-[4-(6-fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl]ethyl}-6,7,8,9-tetrahydro-2-methyl-4H-pyrido[1,2-a]pyrimid-in-4-one
EU-0101074
3-{2-[4-(6-Fluoro-benzo[d]isoxazol-3-yl)-piperidin-1-yl]-ethyl}-2-methyl-6,7,8,9-tetrahydro-pyrido[1,2-a]pyrimidin-4-one (risperidone)
HMS3373M18
4767-EP2275423A1
J10290
4767-EP2308875A1
MCULE-4027175053
NCGC00015883-02
AB00514010-12
NCGC00094352-01
ANW-42874
1026102-43-1
BIDD:GT0262
3-[2-[-4-(6-fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl]ethyl]-6,7,8,9-tetrahydro-2-methyl-4H-pyrido[1,2-a]pyrimidin-4-one
Risperidone [USAN:BAN:INN]
Risperidone, placebo
RTR-001241
Spiron
Tox21_110253_1
NSC759895
R 62 766
R-64766
Risperdal (TN)
3-[2-[4-(6 fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl]ethyl]-6,7,8,9-tetrahydro-2-methyl-4H-pyrido[1,2-a]pyrimidin-4-one
CHEMBL85
3-[2-[4-(6-fluoro-1,2-benzisoxazol-3-yl)piperidin-1-yl]ethyl]-2-methyl-6,7,8,9-tetrahydro-4H-pyrido[1,2-a]pyrimidin-4-one
DB00734
3-{2-[4-(6-Fluoro-1,2-benzisoxazol-3-yl)piperidino]ethyl}-6,7,8,9-tetrahydro-2-methylpyrido[1,2-a]pyrimidin-4-one
HMS1571M19
3-{2-[4-(6-fluorobenzo[d]isoxazol-3-yl)-piperidin-1-yl]-ethyl}-2-methyl-6,7,8,9-tetrahydro-4H-pyrido[1,2-a]pyrimidin-4-one
HMS3715M19
4767-EP2298731A1
KS-1106
4H-Pyrido(1,2-a)pyrimidin-4-one, 3-(2-(4-(6-fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl)ethyl)-6,7,8,9-tetrahydro-2-methyl-
MLS001165758
A801409
NCGC00015883-05
AC-1306
BC206675
2-{2-[4-(6-Fluoro-benzo[d]isoxazol-3-yl)-piperidin-1-yl]-ethyl}-6,7,8,8a-tetrahydro-5H-naphthalen-1-one
BRN 4891881
Risperidone (RIS)
Risperidone, 99%
Risperidonum
SC-16313
SR-01000075399-8
NCGC00094352-02
US8802672, Risperidone
Prestwick0_001029
R 64,766
R64766
risperdone
CAS-106266-06-2
3-[2-[4-(6-Fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl]-6,7,8,9-tetrahydro-2-methyl-4H-pyrido [1,2-a] pyrimidin-4-one
CR0023
3-[2-[4-(6-fluoro-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl]-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one
DSSTox_RID_80740
3-{2-[4-(6-FLUORO-BENZO[D]ISOXAZOL-3-YL)-PIPERIDIN-1-YL]-ETHYL}-2-METHYL-6,7,8,9-TETRAHYDRO-PYRIDO[1,2-A]PYRIMIDIN-4-ONE
HMS2098M19
4767-EP2272841A1
I01-1156
4767-EP2308867A2
Lopac-R-118
4H-Pyrido[1,2-a]pyrimidin-4-one, 3-[2-[4-(6-fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl]ethyl]-6,7,8,9-tetrahydro-2-methyl-
NC00180
AB00514010-09
NCGC00015883-08
AKOS005577302
(risperidone)3-{2-[4-(6-Fluoro-benzo[d]isoxazol-3-yl)-piperidin-1-yl]-ethyl}-2-methyl-6,7,8,9-tetrahydro-pyrido[1,2-a]pyrimidin-4-one
Belivon
3-(2-(4-(6-fluorobenzo[d]isoxazol-3-yl)piperidin-1-yl)ethyl)-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one
Risperidone for system suitability, European Pharmacopoeia (EP) Reference Standard
Risperidone, European Pharmacopoeia (EP) Reference Standard
Rispolept
SMR000466323
TL8000230
NP-202
Zophrenal
PYR332
R-64,766
Rispen
Risperidona
CHEBI:8871
3-[2-[4-(6-fluoro-1,2-benzisoxazol-3-yl)-1-piperidiny]ethyl]-6,7,8,9-tetrahydro-2-methyl-4H-pyrido[1,2-a]pyrimidin-4-one
D01AZG
3-{2-[4-(6-fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl]ethyl}-6,7,8,9-tetrahydro-2-methyl-4H-pyrido[1,2-a]pyrimidin-4-one
FT-0631037
3-{2-[4-(6-Fluoro-benzo[d]isoxazol-3-yl)-piperidin-1-yl]-ethyl}-2-methyl-6,7,8,9-tetrahydro-pyrido[1,2-a]pyrimidin-4-one(risperidone)
HMS3393H07
4767-EP2280008A2
Jsp000573
4767-EP2316836A1
MFCD00274576
66R062
NCGC00015883-03
AB00514010_13
Apexidone
106266-06-2
BRD-K53857191-001-04-5
3-[2-[4(6-fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl]ethyl]-6,7,8,9-tetrahydro-2-methyl-4H-pyrido[1,2-a]pyrimidin-4-one
Risperidone (JAN/USAN/INN)
Risperidone [USAN:USP:INN:BAN]
Risperidone, United States Pharmacopeia (USP) Reference Standard
s1615
SR-01000075399
TR-001241
PB26023
R 64 766
R0087
Risperdal Consta
3-[2-[4-(6-fluoranyl-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl]-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one
Consta, Risperdal
3-[2-[4-(6-fluoro-1,2-benzoxazol-3-yl)-1-piperidinyl]ethyl]-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one
DSSTox_CID_25193
3-{2-[4-(6-fluoro-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl}-2-methyl-4H,6H,7H,8H,9H-pyrido[1,2-a]pyrimidin-4-one
HMS2051H07
4767-EP2269990A1
HSDB 7580
4767-EP2298776A1
L000510
4H-Pyrido(1,2-a)pyrimidin-4-one, 6,7,8,9-tetrahydro-3-(2-(4-(6-fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl)ethyl)-2-methyl-
MLS001424081
AB0004623
NCGC00015883-06
AC1L1JJU
BCP08161
3-(2-(4-(6-Fluoro-1,2-benzisoxazol-3-yl)piperidino)ethyl)-6,7,8,9-tetrahydro-2-methyl-4H-pyrido(1,2-a)pyrimidin-4-one
C-21900
Risperidone (Risperdal)
Risperidone, >=98% (HPLC), powder
Risperidonum [Latin]
SCHEMBL27911
STK646402
NCGC00094352-03
Z1522566617
Prestwick1_001029
R 64766
RAPZEAPATHNIPO-UHFFFAOYSA-N
Risperidal
CC-34268
3-[2-[4-(6-fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl]-ethyl]-6,7,8,9-tetrahydro-2-methyl-4H-pyrido[1,2-A]pyrimidin-4-one
CS-1619
3-[2-[4-(6-fluoro-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl]-2-methyl-6,7,8,9-tetrahydropyrido[2,1-b]pyrimidin-4-one
DTXSID8045193
3-{2-[4-(6-Fluoro-benzo[d]isoxazol-3-yl)-piperidin-1-yl]-ethyl}-2-methyl-6,7,8,9-tetrahydro-pyrido[1,2-a]pyrimidin-4-one (Resperidone)
HMS2233O11
4767-EP2275420A1
J-001555
4767-EP2308870A2
LS-134196
5-[2-(4-Benzo[d]isothiazol-3-yl-piperazin-1-yl)-ethyl]-6-methyl-1,3-dihydro-indol-2-one(Norastemizole)
NCGC00015883-01
AB00514010-11
NCGC00015883-11
AN-15065
BG0309
3-?[2-?[4-?(6-?fluoro-?1,?2-?benzisoxazol-?3-?yl)?-?1-?piperidinyl]?ethyl]?-?2,?3,?6,?7,?8,?9-?hexahydro-?2-?methyl-4H-?Pyrido[1,?2-?a]?pyrimidin-?4-?one [ Show all ] |
|---|


![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218