

You can:
| Name | 5-hydroxytryptamine receptor 1A |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Htr1a |
| Synonym | 5-HT1A receptor 5-hydroxytryptamine (serotonin) receptor 1A, G protein-coupled 5-HT1A ADRB2RL1 ADRBRL1 [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 422 |
| Amino acid sequence | MDVFSFGQGNNTTASQEPFGTGGNVTSISDVTFSYQVITSLLLGTLIFCAVLGNACVVAAIALERSLQNVANYLIGSLAVTDLMVSVLVLPMAALYQVLNKWTLGQVTCDLFIALDVLCCTSSILHLCAIALDRYWAITDPIDYVNKRTPRRAAALISLTWLIGFLISIPPMLGWRTPEDRSDPDACTISKDHGYTIYSTFGAFYIPLLLMLVLYGRIFRAARFRIRKTVRKVEKKGAGTSLGTSSAPPPKKSLNGQPGSGDWRRCAENRAVGTPCTNGAVRQGDDEATLEVIEVHRVGNSKEHLPLPSESGSNSYAPACLERKNERNAEAKRKMALARERKTVKTLGIIMGTFILCWLPFFIVALVLPFCESSCHMPALLGAIINWLGYSNSLLNPVIYAYFNKDFQNAFKKIIKCKFCRR |
| UniProt | P19327 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL273 |
| IUPHAR | 1 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 41 | 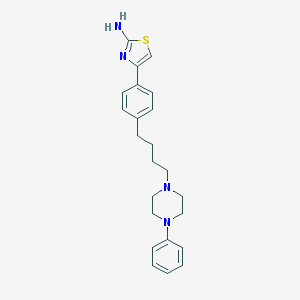 CHEMBL171316 CHEMBL171316 | C23H28N4S | 392.565 | 5 / 1 | 4.9 | Yes |
| 557308 | 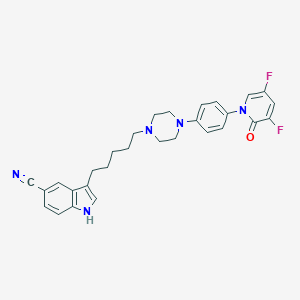 3-(5-(4-(4-(3,5-difluoro-2-oxopyridin-1(2H)-yl)phenyl)piperazin-1-yl)pentyl)-1H-indole-5-carbonitrile 3-(5-(4-(4-(3,5-difluoro-2-oxopyridin-1(2H)-yl)phenyl)piperazin-1-yl)pentyl)-1H-indole-5-carbonitrile | C29H29F2N5O | 501.582 | 6 / 1 | 5.1 | No |
| 53 | 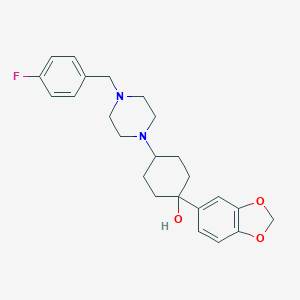 CHEMBL100149 CHEMBL100149 | C24H29FN2O3 | 412.505 | 6 / 1 | 3.4 | Yes |
| 555477 | 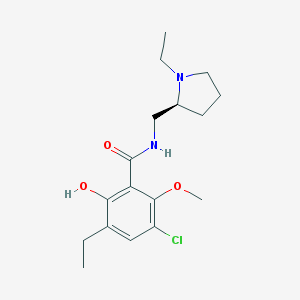 Eticlopride Eticlopride | C17H25ClN2O3 | 340.848 | 4 / 2 | 3.1 | Yes |
| 273 | 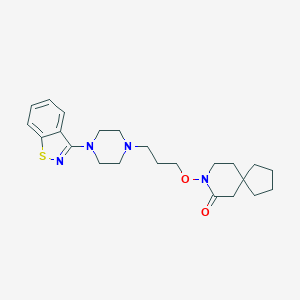 CHEMBL269176 CHEMBL269176 | C23H32N4O2S | 428.595 | 6 / 0 | 4.6 | Yes |
| 275 | 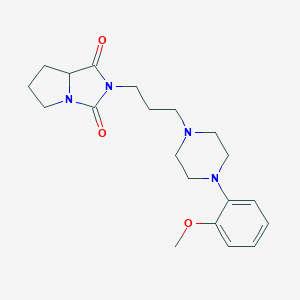 2-{3-[4-(2-methoxyphenyl)-1-piperazinyl]propyl}tetrahydro-1H-pyrrolo[1,2-c]imidazole-1,3(2H)-dione 2-{3-[4-(2-methoxyphenyl)-1-piperazinyl]propyl}tetrahydro-1H-pyrrolo[1,2-c]imidazole-1,3(2H)-dione | C20H28N4O3 | 372.469 | 5 / 0 | 1.8 | Yes |
| 292 | 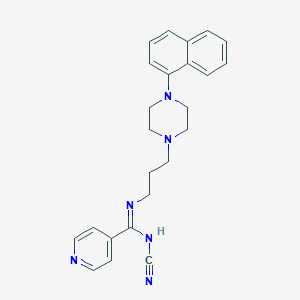 CHEMBL1783352 CHEMBL1783352 | C24H26N6 | 398.514 | 5 / 1 | 3.5 | Yes |
| 333 | 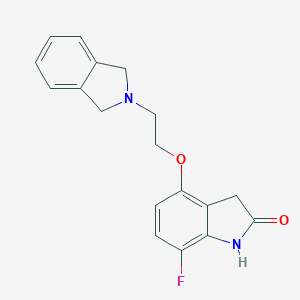 CHEMBL300316 CHEMBL300316 | C18H17FN2O2 | 312.344 | 4 / 1 | 1.9 | Yes |
| 547905 | 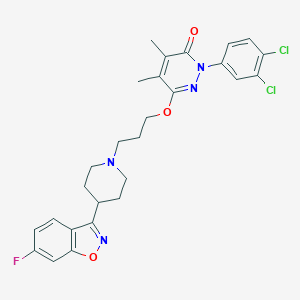 CHEMBL3883388 CHEMBL3883388 | C27H27Cl2FN4O3 | 545.436 | 7 / 0 | 6.0 | No |
| 623 | 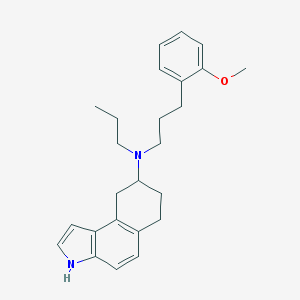 CHEMBL306197 CHEMBL306197 | C25H32N2O | 376.544 | 2 / 1 | 6.0 | No |
| 709 | 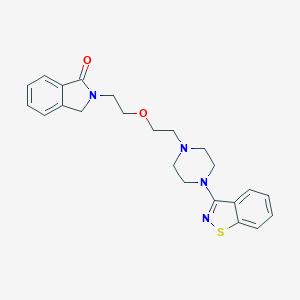 CHEMBL288450 CHEMBL288450 | C23H26N4O2S | 422.547 | 6 / 0 | 3.2 | Yes |
| 891 | 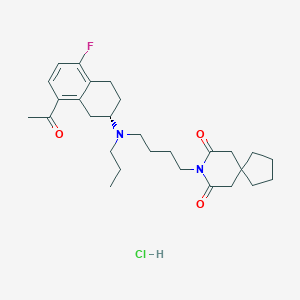 CHEMBL540064 CHEMBL540064 | C28H40ClFN2O3 | 507.087 | 5 / 1 | N/A | No |
| 892 | 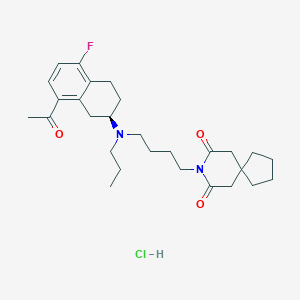 CHEMBL542474 CHEMBL542474 | C28H40ClFN2O3 | 507.087 | 5 / 1 | N/A | No |
| 961 | 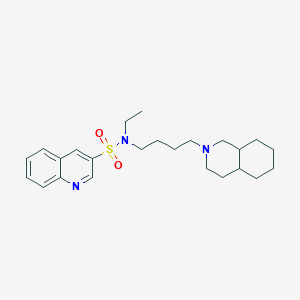 CHEMBL1917358 CHEMBL1917358 | C24H35N3O2S | 429.623 | 5 / 0 | 4.9 | Yes |
| 1190 | 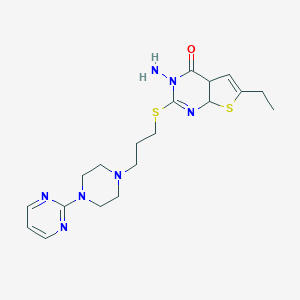 CID 44578448 CID 44578448 | C19H27N7OS2 | 433.593 | 9 / 1 | 1.7 | Yes |
| 1191 | 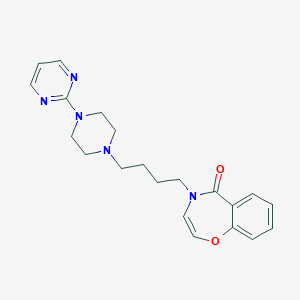 CHEMBL154145 CHEMBL154145 | C21H25N5O2 | 379.464 | 6 / 0 | 2.1 | Yes |
| 1306 | 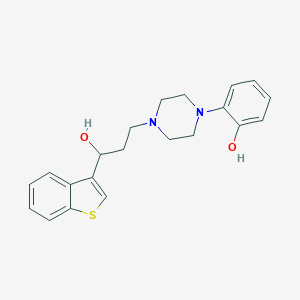 CHEMBL135890 CHEMBL135890 | C21H24N2O2S | 368.495 | 5 / 2 | 3.7 | Yes |
| 1411 | 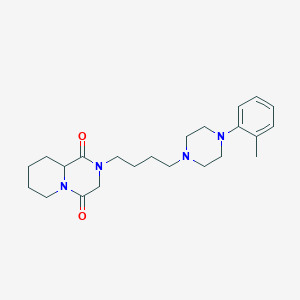 CHEMBL58702 CHEMBL58702 | C23H34N4O2 | 398.551 | 4 / 0 | 2.6 | Yes |
| 441724 | 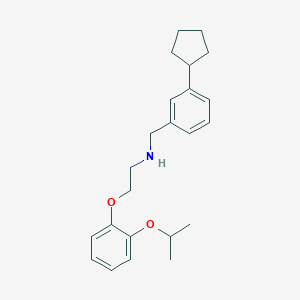 CHEMBL413777 CHEMBL413777 | C23H31NO2 | 353.506 | 3 / 1 | 5.7 | No |
| 1527 | 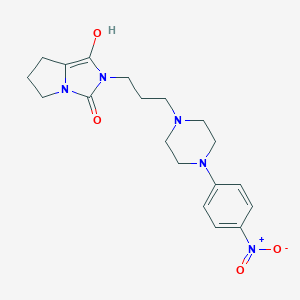 BDBM50054356 BDBM50054356 | C19H25N5O4 | 387.44 | 6 / 1 | 1.6 | Yes |
| 1541 | 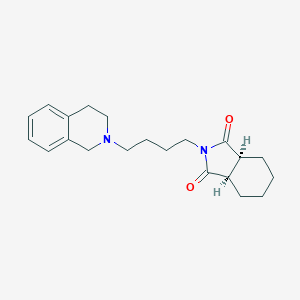 CHEMBL146989 CHEMBL146989 | C21H28N2O2 | 340.467 | 3 / 0 | 3.1 | Yes |
| 1542 | 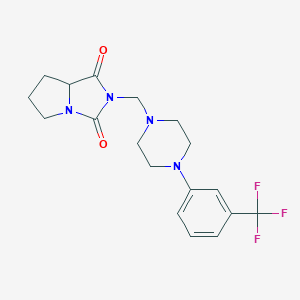 Tetrahydro-2-[[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]methyl]-1H-pyrrolo[1,2-c]imidazole-1,3(2H)-dione Tetrahydro-2-[[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]methyl]-1H-pyrrolo[1,2-c]imidazole-1,3(2H)-dione | C18H21F3N4O2 | 382.387 | 7 / 0 | 2.5 | Yes |
| 1741 | 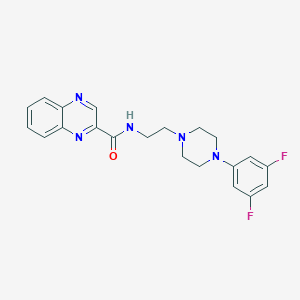 CHEMBL3264371 CHEMBL3264371 | C21H21F2N5O | 397.43 | 7 / 1 | 2.6 | Yes |
| 1795 | 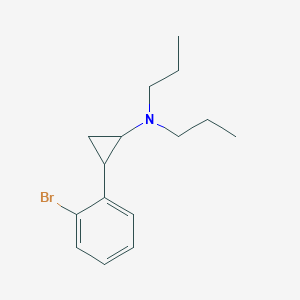 CHEMBL542094 CHEMBL542094 | C15H22BrN | 296.252 | 1 / 0 | 4.7 | Yes |
| 1963 | 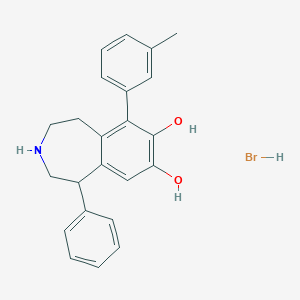 CHEMBL484356 CHEMBL484356 | C23H24BrNO2 | 426.354 | 3 / 4 | N/A | N/A |
| 1966 | 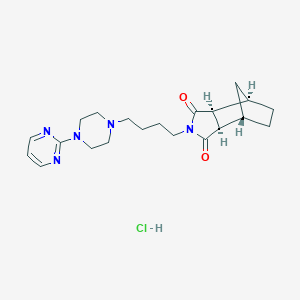 TANDOSPIRONE HYDROCHLORIDE TANDOSPIRONE HYDROCHLORIDE | C21H30ClN5O2 | 419.954 | 6 / 1 | N/A | N/A |
| 521503 | 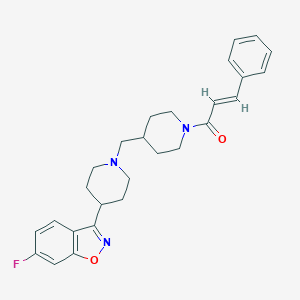 CHEMBL3741516 CHEMBL3741516 | C27H30FN3O2 | 447.554 | 5 / 0 | 4.7 | Yes |
| 1979 | 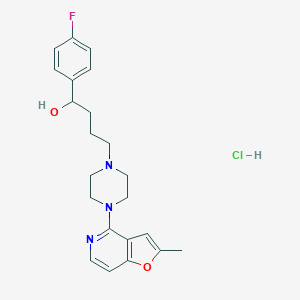 CHEMBL557995 CHEMBL557995 | C22H27ClFN3O2 | 419.925 | 6 / 2 | N/A | N/A |
| 1993 | 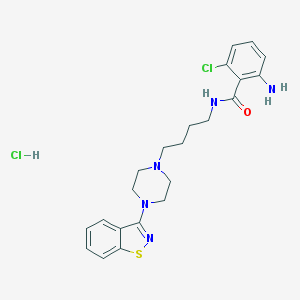 CHEMBL545372 CHEMBL545372 | C22H27Cl2N5OS | 480.452 | 6 / 3 | N/A | N/A |
| 2063 | 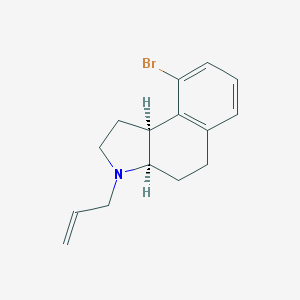 CHEMBL164195 CHEMBL164195 | C15H18BrN | 292.22 | 1 / 0 | 3.9 | Yes |
| 2105 | 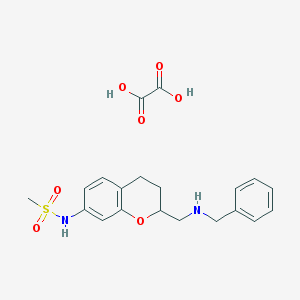 CHEMBL337696 CHEMBL337696 | C20H24N2O7S | 436.479 | 9 / 4 | N/A | N/A |
| 2194 | 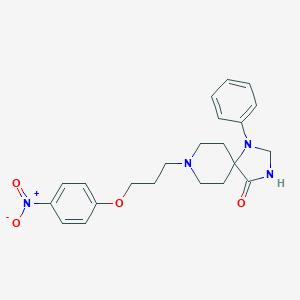 CHEMBL309992 CHEMBL309992 | C22H26N4O4 | 410.474 | 6 / 1 | 3.1 | Yes |
| 2251 | 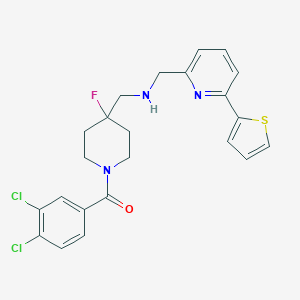 CHEMBL288244 CHEMBL288244 | C23H22Cl2FN3OS | 478.407 | 5 / 1 | 4.8 | Yes |
| 2977 | 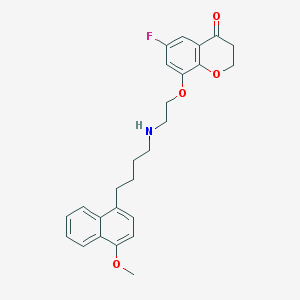 CHEMBL317532 CHEMBL317532 | C26H28FNO4 | 437.511 | 6 / 1 | 4.9 | Yes |
| 3119 | 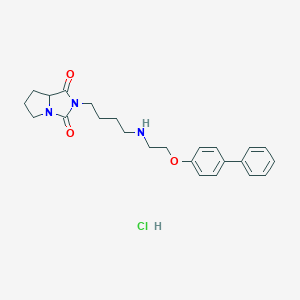 CHEMBL2429880 CHEMBL2429880 | C24H30ClN3O3 | 443.972 | 4 / 2 | N/A | N/A |
| 3350 | 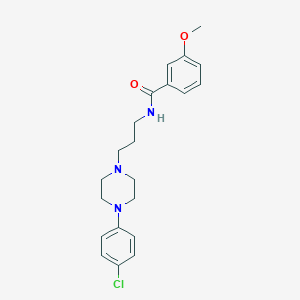 CHEMBL92713 CHEMBL92713 | C21H26ClN3O2 | 387.908 | 4 / 1 | 3.8 | Yes |
| 3380 | 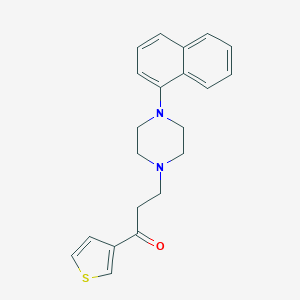 CHEMBL135295 CHEMBL135295 | C21H22N2OS | 350.48 | 4 / 0 | 4.1 | Yes |
| 3412 | 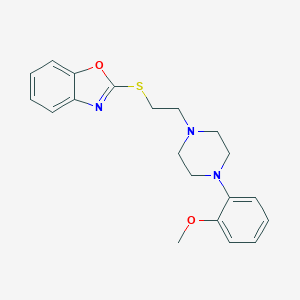 CHEMBL476363 CHEMBL476363 | C20H23N3O2S | 369.483 | 6 / 0 | 4.2 | Yes |
| 3488 | 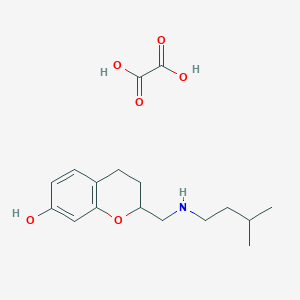 CHEMBL134843 CHEMBL134843 | C17H25NO6 | 339.388 | 7 / 4 | N/A | N/A |
| 3660 | 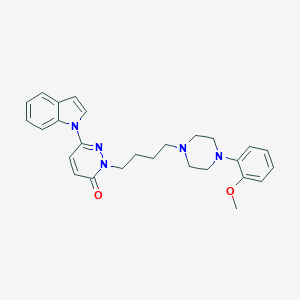 CHEMBL420718 CHEMBL420718 | C27H31N5O2 | 457.578 | 5 / 0 | 3.9 | Yes |
| 3688 | 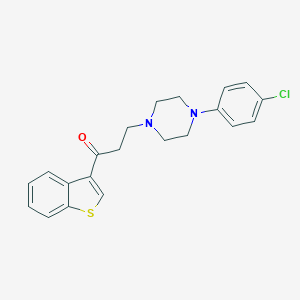 CHEMBL137185 CHEMBL137185 | C21H21ClN2OS | 384.922 | 4 / 0 | 5.0 | Yes |
| 3724 | 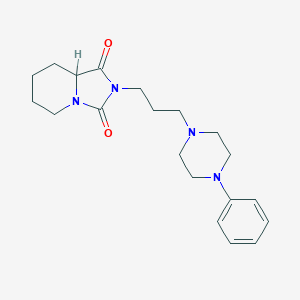 CID 10665976 CID 10665976 | C20H28N4O2 | 356.47 | 4 / 0 | 2.2 | Yes |
| 3754 | 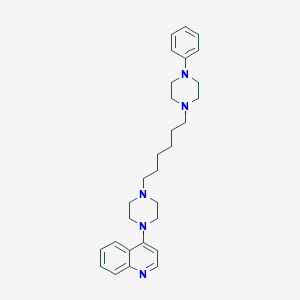 CHEMBL1173017 CHEMBL1173017 | C29H39N5 | 457.666 | 5 / 0 | 5.2 | No |
| 3765 | 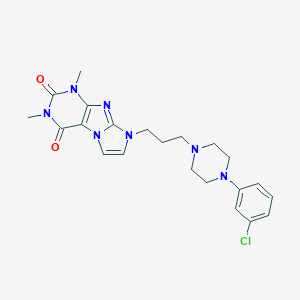 CHEMBL566890 CHEMBL566890 | C22H26ClN7O2 | 455.947 | 5 / 0 | 3.0 | Yes |
| 3774 | 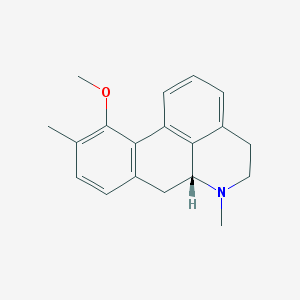 CHEMBL332178 CHEMBL332178 | C19H21NO | 279.383 | 2 / 0 | 3.3 | Yes |
| 3782 | 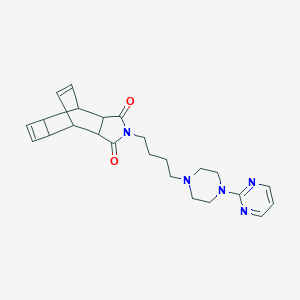 CHEMBL1204094 CHEMBL1204094 | C24H29N5O2 | 419.529 | 6 / 0 | 1.4 | Yes |
| 3792 | 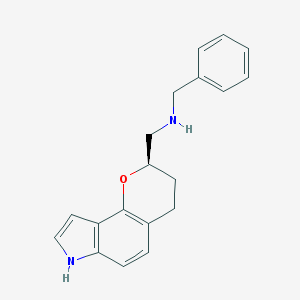 CHEMBL60185 CHEMBL60185 | C19H20N2O | 292.382 | 2 / 2 | 3.5 | Yes |
| 3934 | 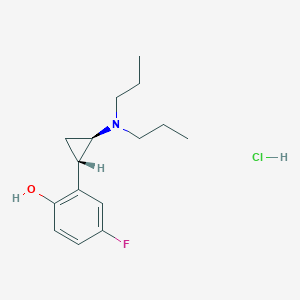 CHEMBL545643 CHEMBL545643 | C15H23ClFNO | 287.803 | 3 / 2 | N/A | N/A |
| 3935 | 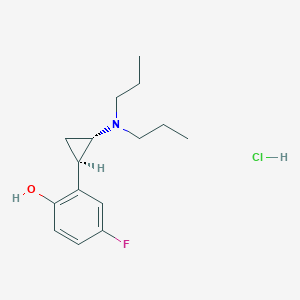 CHEMBL542589 CHEMBL542589 | C15H23ClFNO | 287.803 | 3 / 2 | N/A | N/A |
| 3936 | 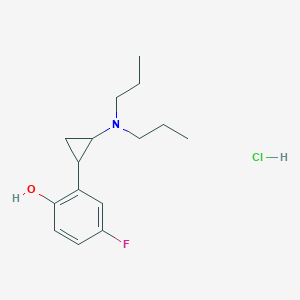 CHEMBL539497 CHEMBL539497 | C15H23ClFNO | 287.803 | 3 / 2 | N/A | N/A |
| 521561 | 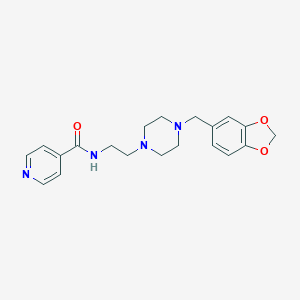 CHEMBL3764080 CHEMBL3764080 | C20H24N4O3 | 368.437 | 6 / 1 | 1.3 | Yes |
| 4129 | 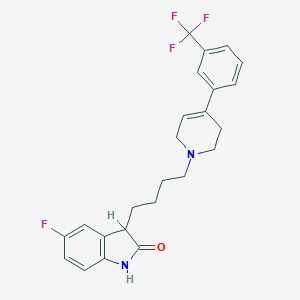 CHEMBL1836792 CHEMBL1836792 | C24H24F4N2O | 432.463 | 6 / 1 | 4.8 | Yes |
| 4152 | 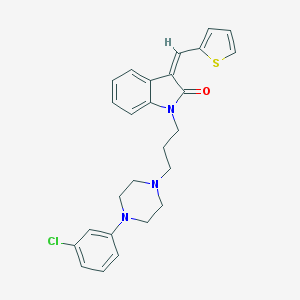 CHEMBL282255 CHEMBL282255 | C26H26ClN3OS | 464.024 | 4 / 0 | 5.4 | No |
| 4154 | 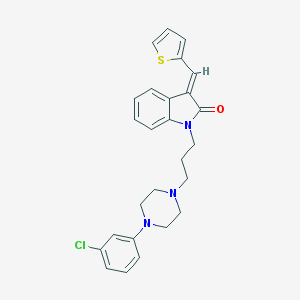 CHEMBL21150 CHEMBL21150 | C26H26ClN3OS | 464.024 | 4 / 0 | 5.4 | No |
| 4221 | 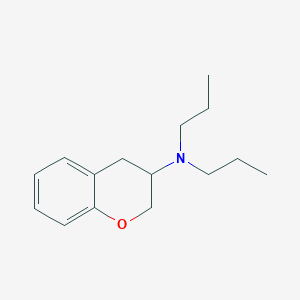 CHEMBL25984 CHEMBL25984 | C15H23NO | 233.355 | 2 / 0 | 3.8 | Yes |
| 4409 | 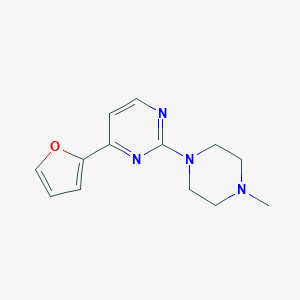 CHEMBL295689 CHEMBL295689 | C13H16N4O | 244.298 | 5 / 0 | 1.2 | Yes |
| 521575 | 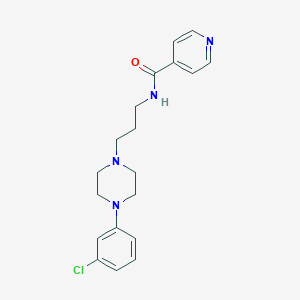 CHEMBL3765273 CHEMBL3765273 | C19H23ClN4O | 358.87 | 4 / 1 | 2.7 | Yes |
| 4497 | 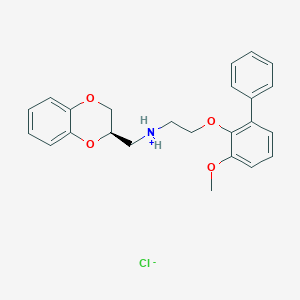 CID 44417700 CID 44417700 | C24H26ClNO4 | 427.925 | 5 / 1 | N/A | N/A |
| 4501 | 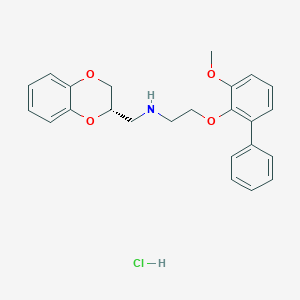 CHEMBL215151 CHEMBL215151 | C24H26ClNO4 | 427.925 | 5 / 2 | N/A | N/A |
| 4507 | 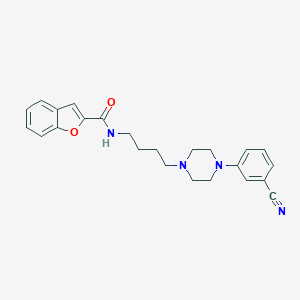 CHEMBL497604 CHEMBL497604 | C24H26N4O2 | 402.498 | 5 / 1 | 4.0 | Yes |
| 441886 | 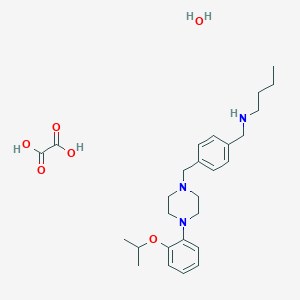 CHEMBL3392290 CHEMBL3392290 | C27H41N3O6 | 503.64 | 9 / 4 | N/A | No |
| 4598 | 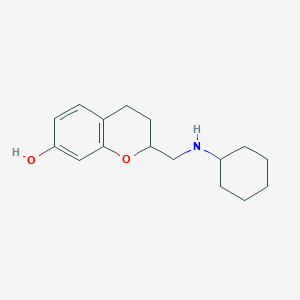 CHEMBL135011 CHEMBL135011 | C16H23NO2 | 261.365 | 3 / 2 | 3.4 | Yes |
| 547946 | 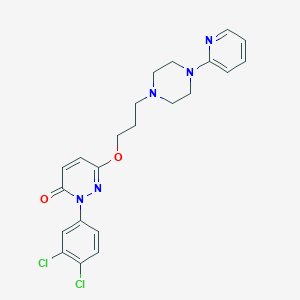 CHEMBL3883429 CHEMBL3883429 | C22H23Cl2N5O2 | 460.359 | 6 / 0 | 3.9 | Yes |
| 4666 | 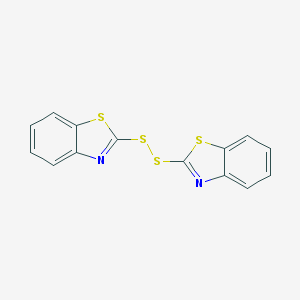 120-78-5 120-78-5 | C14H8N2S4 | 332.472 | 6 / 0 | 5.6 | No |
| 4695 |  CHEMBL1202520 CHEMBL1202520 | C26H31N5O2 | 445.567 | 6 / 0 | 1.9 | Yes |
| 4949 |  CHEMBL3233865 CHEMBL3233865 | C30H32N2O4 | 484.596 | 6 / 0 | 5.6 | No |
| 4973 |  CHEMBL50188 CHEMBL50188 | C25H36N2O4 | 428.573 | 5 / 0 | 4.2 | Yes |
| 4979 |  CHEMBL190525 CHEMBL190525 | C26H29N5O3 | 459.55 | 5 / 2 | 3.4 | Yes |
| 5228 |  CHEMBL1203171 CHEMBL1203171 | C19H24N2OS | 328.474 | 3 / 2 | 4.1 | Yes |
| 5363 |  CHEMBL41536 CHEMBL41536 | C24H26N4O2S | 434.558 | 6 / 0 | 4.3 | Yes |
| 5410 |  CHEMBL403844 CHEMBL403844 | C19H19N | 261.368 | 1 / 0 | 3.7 | Yes |
| 5434 |  CHEMBL283528 CHEMBL283528 | C19H28ClN3O2 | 365.902 | 4 / 1 | 3.5 | Yes |
| 5436 |  CHEMBL279729 CHEMBL279729 | C19H28ClN3O2 | 365.902 | 4 / 1 | 3.5 | Yes |
| 5546 |  CHEMBL50722 CHEMBL50722 | C25H36N2O5 | 444.572 | 6 / 0 | 4.3 | Yes |
| 5556 |  CHEMBL23912 CHEMBL23912 | C21H28N4O2 | 368.481 | 5 / 1 | 2.4 | Yes |
| 5578 |  CHEMBL3144354 CHEMBL3144354 | C28H29N3O4S | 503.617 | 6 / 1 | 4.0 | No |
| 5622 |  CHEMBL1836788 CHEMBL1836788 | C21H26N4O | 350.466 | 4 / 1 | 2.8 | Yes |
| 5697 |  CHEMBL544909 CHEMBL544909 | C23H29N5OS | 423.579 | 6 / 2 | 4.5 | Yes |
| 5714 | 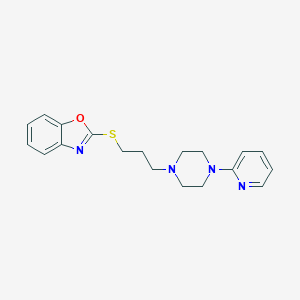 CHEMBL517962 CHEMBL517962 | C19H22N4OS | 354.472 | 6 / 0 | 3.9 | Yes |
| 5766 | 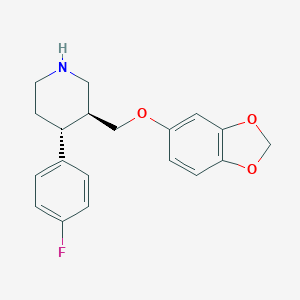 paroxetine paroxetine | C19H20FNO3 | 329.371 | 5 / 1 | 3.5 | Yes |
| 5813 | 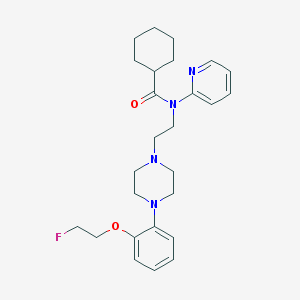 CHEMBL137298 CHEMBL137298 | C26H35FN4O2 | 454.59 | 6 / 0 | 4.6 | Yes |
| 5850 | 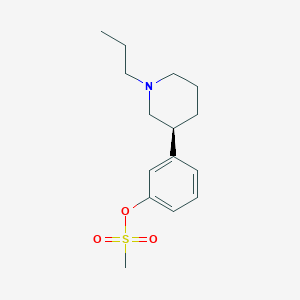 CHEMBL93594 CHEMBL93594 | C15H23NO3S | 297.413 | 4 / 0 | 2.9 | Yes |
| 5918 | 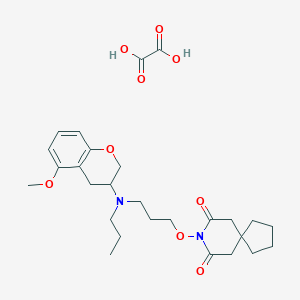 CHEMBL50722 CHEMBL50722 | C27H38N2O9 | 534.606 | 10 / 2 | N/A | No |
| 6337 | 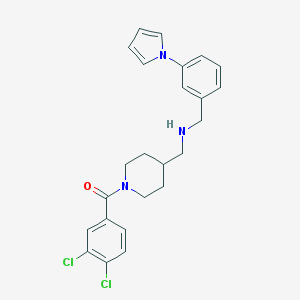 CHEMBL150109 CHEMBL150109 | C24H25Cl2N3O | 442.384 | 2 / 1 | 4.8 | Yes |
| 6363 | 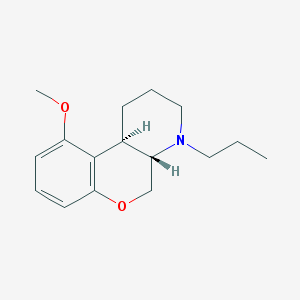 CHEMBL163281 CHEMBL163281 | C16H23NO2 | 261.365 | 3 / 0 | 3.2 | Yes |
| 6534 | 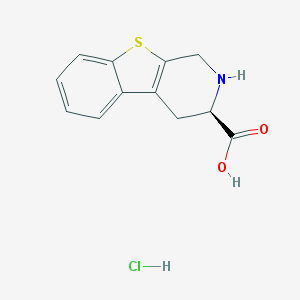 CHEMBL1203203 CHEMBL1203203 | C12H12ClNO2S | 269.743 | 4 / 3 | N/A | N/A |
| 6693 | 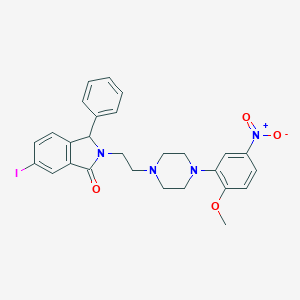 CHEMBL43209 CHEMBL43209 | C27H27IN4O4 | 598.441 | 6 / 0 | 4.7 | No |
| 441969 | 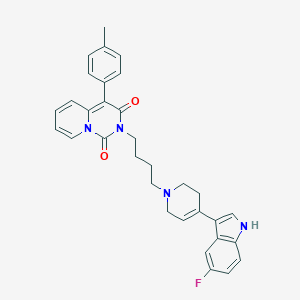 CHEMBL3397077 CHEMBL3397077 | C32H31FN4O2 | 522.624 | 4 / 1 | 5.2 | No |
| 7247 | 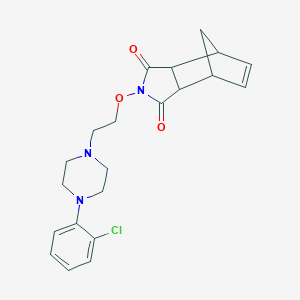 CHEMBL426813 CHEMBL426813 | C21H24ClN3O3 | 401.891 | 5 / 0 | 2.6 | Yes |
| 7279 | 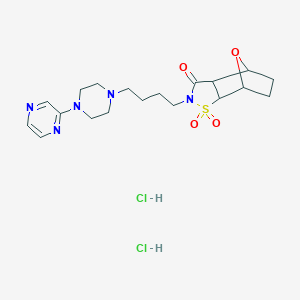 CHEMBL3215745 CHEMBL3215745 | C19H29Cl2N5O4S | 494.432 | 8 / 2 | N/A | N/A |
| 7333 | 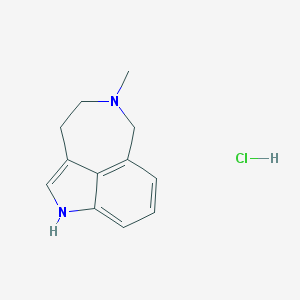 CHEMBL1203613 CHEMBL1203613 | C12H15ClN2 | 222.716 | 1 / 2 | N/A | N/A |
| 7378 | 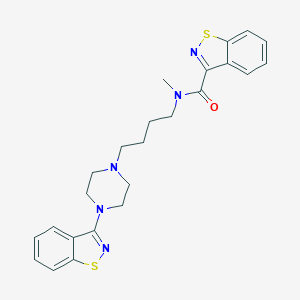 CHEMBL75670 CHEMBL75670 | C24H27N5OS2 | 465.634 | 7 / 0 | 5.2 | No |
| 7408 | 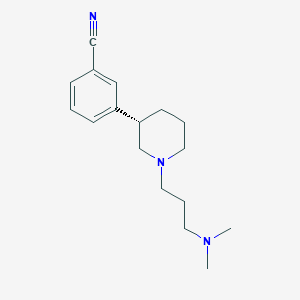 CHEMBL327836 CHEMBL327836 | C17H25N3 | 271.408 | 3 / 0 | 2.6 | Yes |
| 7444 | 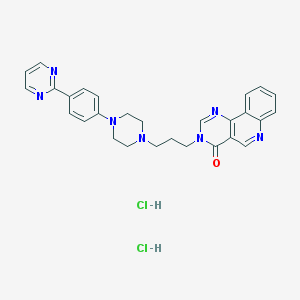 CHEMBL3215842 CHEMBL3215842 | C28H29Cl2N7O | 550.488 | 7 / 2 | N/A | No |
| 7573 | 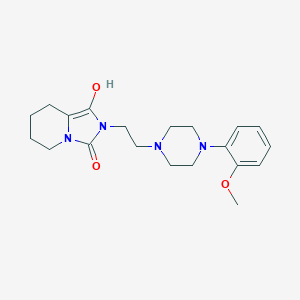 BDBM50054361 BDBM50054361 | C20H28N4O3 | 372.469 | 5 / 1 | 1.7 | Yes |
| 7686 | 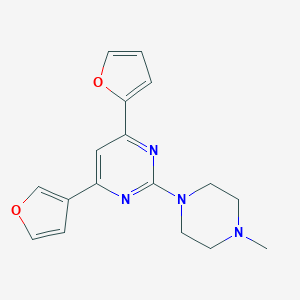 CHEMBL42061 CHEMBL42061 | C17H18N4O2 | 310.357 | 6 / 0 | 2.0 | Yes |
| 7693 | 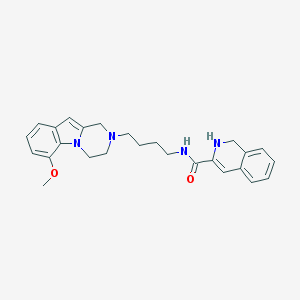 CID 44581146 CID 44581146 | C26H30N4O2 | 430.552 | 4 / 2 | 3.5 | Yes |
| 7807 | 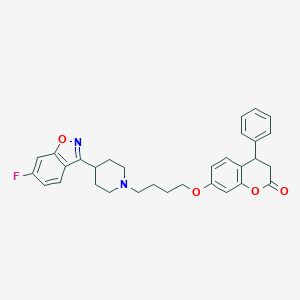 CHEMBL2387242 CHEMBL2387242 | C31H31FN2O4 | 514.597 | 7 / 0 | 6.0 | No |
| 7926 |  CHEMBL3121412 CHEMBL3121412 | C31H30N2O4 | 494.591 | 5 / 2 | N/A | N/A |
| 8094 |  CHEMBL291620 CHEMBL291620 | C18H29NO | 275.436 | 2 / 0 | 4.8 | Yes |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218