

You can:
| Name | G-protein coupled receptor 35 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GPR35 |
| Synonym | G-protein coupled receptor 3 GPR35 KYNA receptor Kynurenic acid receptor |
| Disease | N/A |
| Length | 309 |
| Amino acid sequence | MNGTYNTCGSSDLTWPPAIKLGFYAYLGVLLVLGLLLNSLALWVFCCRMQQWTETRIYMTNLAVADLCLLCTLPFVLHSLRDTSDTPLCQLSQGIYLTNRYMSISLVTAIAVDRYVAVRHPLRARGLRSPRQAAAVCAVLWVLVIGSLVARWLLGIQEGGFCFRSTRHNFNSMAFPLLGFYLPLAVVVFCSLKVVTALAQRPPTDVGQAEATRKAARMVWANLLVFVVCFLPLHVGLTVRLAVGWNACALLETIRRALYITSKLSDANCCLDAICYYYMAKEFQEASALAVAPSAKAHKSQDSLCVTLA |
| UniProt | Q9HC97 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9HC97 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9HC97. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL1293267 |
| IUPHAR | 102 |
| DrugBank | BE0005562 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 3811 | 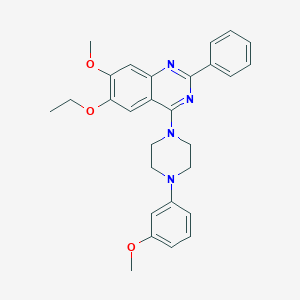 CHEMBL2431236 CHEMBL2431236 | C28H30N4O3 | 470.573 | 7 / 0 | 5.5 | No |
| 4428 | 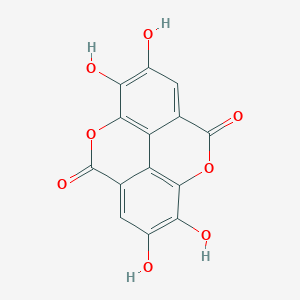 ellagic acid ellagic acid | C14H6O8 | 302.194 | 8 / 4 | 1.1 | Yes |
| 4534 | 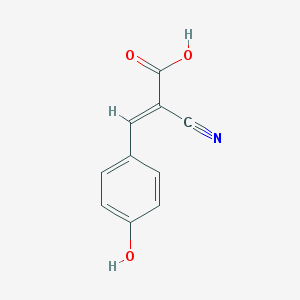 alpha-cyano-4-hydroxycinnamic acid alpha-cyano-4-hydroxycinnamic acid | C10H7NO3 | 189.17 | 4 / 2 | 1.4 | Yes |
| 5317 | 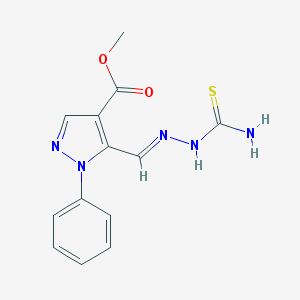 MLS-0437447.0001 MLS-0437447.0001 | C13H13N5O2S | 303.34 | 5 / 2 | 1.4 | Yes |
| 5320 | 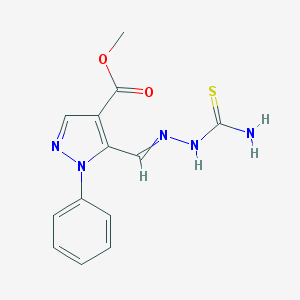 AC1MCK42 AC1MCK42 | C13H13N5O2S | 303.34 | 5 / 2 | 1.4 | Yes |
| 6305 | 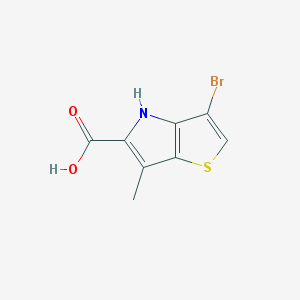 CHEMBL2037465 CHEMBL2037465 | C8H6BrNO2S | 260.105 | 3 / 2 | 2.7 | Yes |
| 442011 | 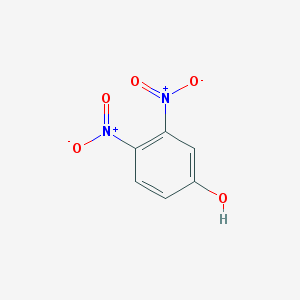 3,4-DINITROPHENOL 3,4-DINITROPHENOL | C6H4N2O5 | 184.107 | 5 / 1 | 1.2 | Yes |
| 9442 | 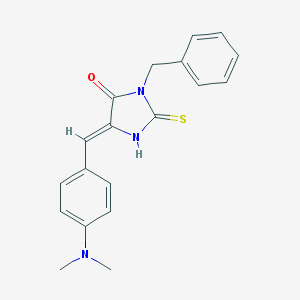 MLS000545777 MLS000545777 | C19H19N3OS | 337.441 | 3 / 1 | 3.5 | Yes |
| 557648 | 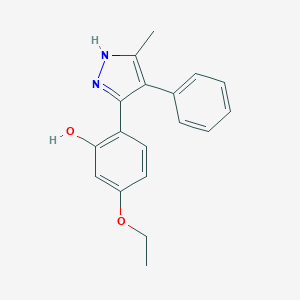 5-ethoxy-2-(5-methyl-4-phenyl-1H-pyrazol-3-yl)phenol 5-ethoxy-2-(5-methyl-4-phenyl-1H-pyrazol-3-yl)phenol | C18H18N2O2 | 294.354 | 3 / 2 | 3.9 | Yes |
| 14703 | 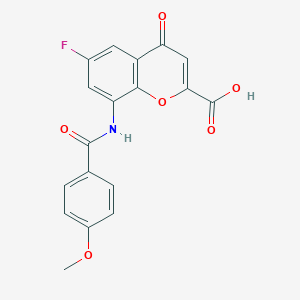 CHEMBL2392162 CHEMBL2392162 | C18H12FNO6 | 357.293 | 7 / 2 | 2.2 | Yes |
| 17294 | 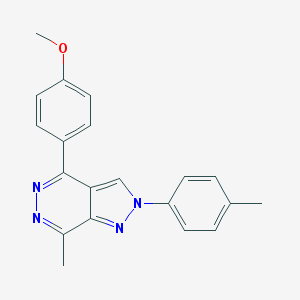 MLS-0437176.0001 MLS-0437176.0001 | C20H18N4O | 330.391 | 4 / 0 | 3.6 | Yes |
| 17523 | 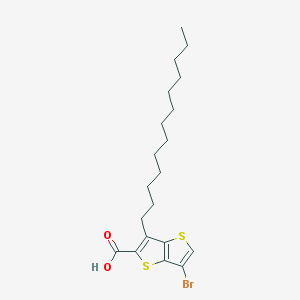 CHEMBL1914591 CHEMBL1914591 | C20H29BrO2S2 | 445.474 | 4 / 1 | 10.0 | No |
| 17812 | 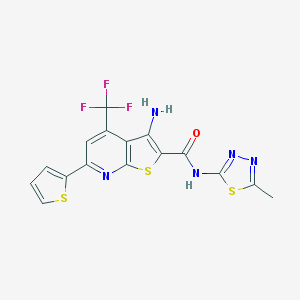 SMR000129696 SMR000129696 | C16H10F3N5OS3 | 441.465 | 11 / 2 | 4.7 | No |
| 19200 | 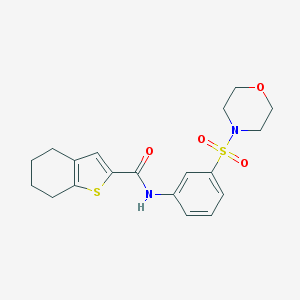 MLS-0435573.0001 MLS-0435573.0001 | C19H22N2O4S2 | 406.515 | 6 / 1 | 2.8 | Yes |
| 21027 | 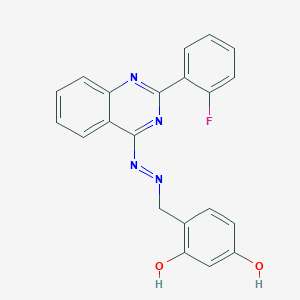 BDBM61566 BDBM61566 | C21H15FN4O2 | 374.375 | 7 / 2 | 4.6 | Yes |
| 22237 | 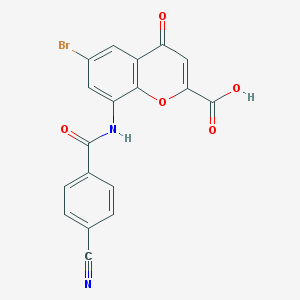 CHEMBL2392173 CHEMBL2392173 | C18H9BrN2O5 | 413.183 | 6 / 2 | 2.5 | Yes |
| 22851 | 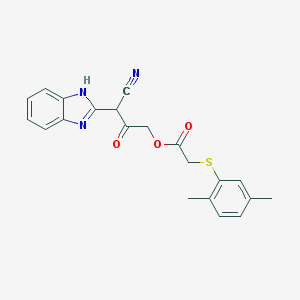 BDBM46570 BDBM46570 | C21H19N3O3S | 393.461 | 6 / 1 | 4.0 | Yes |
| 24621 | 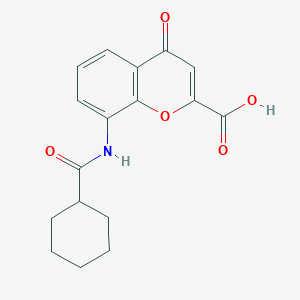 CHEMBL2392148 CHEMBL2392148 | C17H17NO5 | 315.325 | 5 / 2 | 2.5 | Yes |
| 558050 | 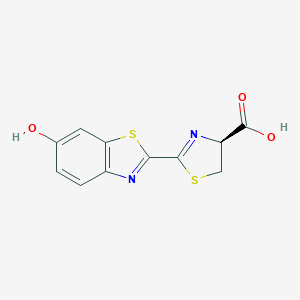 D-Luciferin D-Luciferin | C11H8N2O3S2 | 280.316 | 7 / 2 | 2.2 | Yes |
| 25880 | 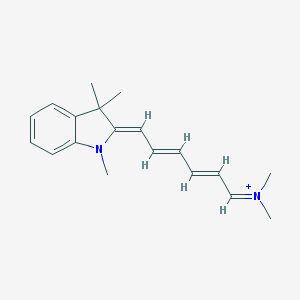 MLS000724457 MLS000724457 | C19H25N2+ | 281.423 | 1 / 0 | 3.9 | Yes |
| 27613 |  MLS-0413566.0001 MLS-0413566.0001 | C19H20N2O7S | 420.436 | 8 / 1 | 1.3 | Yes |
| 28121 |  MLS-0300380.0001 MLS-0300380.0001 | C19H17N5O2S | 379.438 | 5 / 2 | 3.3 | Yes |
| 28125 |  AC1MCK46 AC1MCK46 | C19H17N5O2S | 379.438 | 5 / 2 | 3.3 | Yes |
| 28206 |  ST50920082 ST50920082 | C23H16N4O3S3 | 492.586 | 8 / 2 | 4.3 | Yes |
| 28936 | 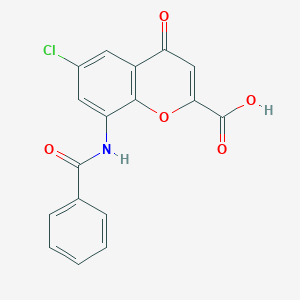 CHEMBL2392164 CHEMBL2392164 | C17H10ClNO5 | 343.719 | 5 / 2 | 2.7 | Yes |
| 35004 | 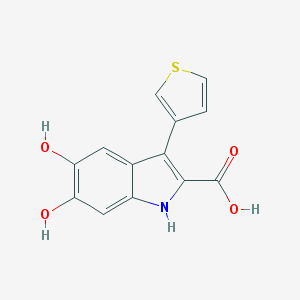 CHEMBL2086422 CHEMBL2086422 | C13H9NO4S | 275.278 | 5 / 4 | 2.5 | Yes |
| 35694 | 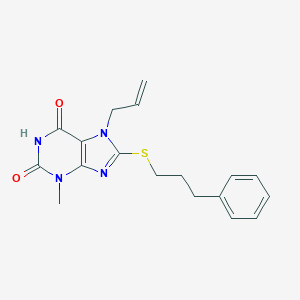 MLS-0061481.0001 MLS-0061481.0001 | C18H20N4O2S | 356.444 | 4 / 1 | 3.3 | Yes |
| 36728 | 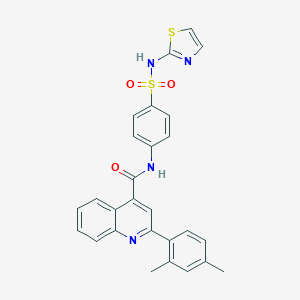 MLS-0203836.0001 MLS-0203836.0001 | C27H22N4O3S2 | 514.618 | 7 / 2 | 5.3 | No |
| 36830 | 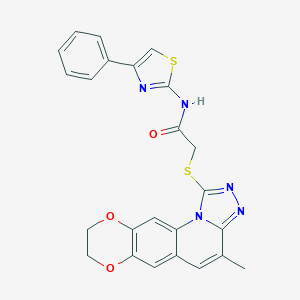 ASN 04482516 ASN 04482516 | C24H19N5O3S2 | 489.568 | 8 / 1 | 5.4 | No |
| 36888 | 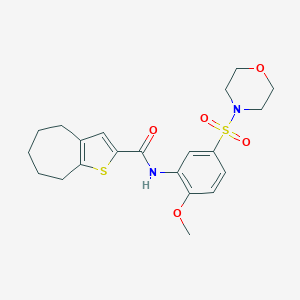 MLS-0435537.0001 MLS-0435537.0001 | C21H26N2O5S2 | 450.568 | 7 / 1 | 3.3 | Yes |
| 38035 | 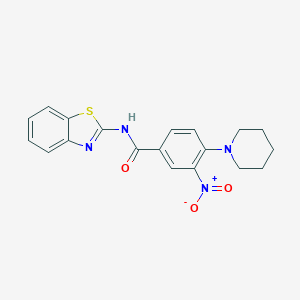 MLS000581649 MLS000581649 | C19H18N4O3S | 382.438 | 6 / 1 | 4.3 | Yes |
| 38511 | 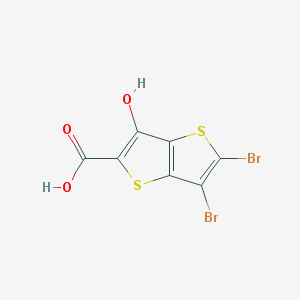 CHEMBL2037456 CHEMBL2037456 | C7H2Br2O3S2 | 358.018 | 5 / 2 | 4.5 | Yes |
| 38861 | 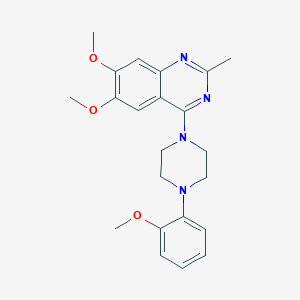 CHEMBL2431113 CHEMBL2431113 | C22H26N4O3 | 394.475 | 7 / 0 | 3.9 | Yes |
| 39229 | 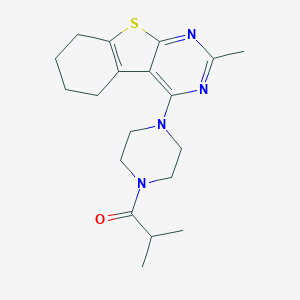 MLS-0437226.0001 MLS-0437226.0001 | C19H26N4OS | 358.504 | 5 / 0 | 4.0 | Yes |
| 39993 | 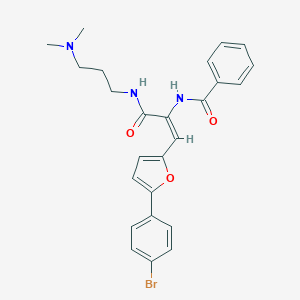 MLS000701097 MLS000701097 | C25H26BrN3O3 | 496.405 | 4 / 2 | 4.5 | Yes |
| 40962 | 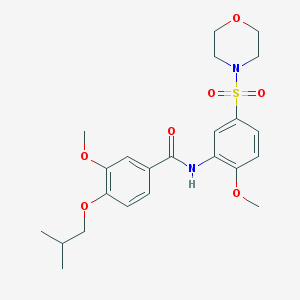 MLS-0413763.0001 MLS-0413763.0001 | C23H30N2O7S | 478.56 | 8 / 1 | 2.8 | Yes |
| 42270 | 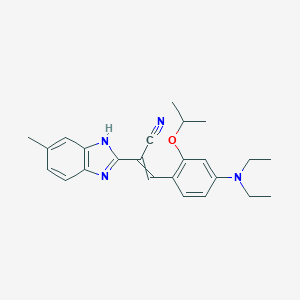 AC1ME79R AC1ME79R | C24H28N4O | 388.515 | 4 / 1 | 5.3 | No |
| 42271 | 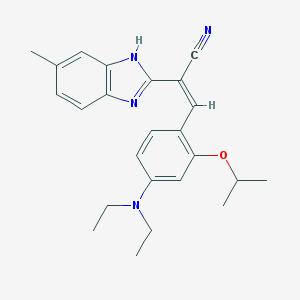 MLS000577677 MLS000577677 | C24H28N4O | 388.515 | 4 / 1 | 5.3 | No |
| 42832 | 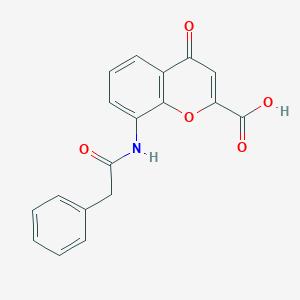 CHEMBL2392150 CHEMBL2392150 | C18H13NO5 | 323.304 | 5 / 2 | 2.1 | Yes |
| 43027 | 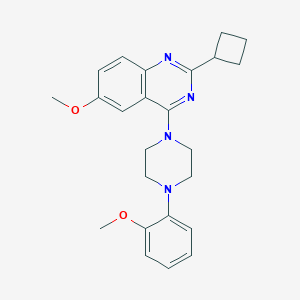 CHEMBL2431125 CHEMBL2431125 | C24H28N4O2 | 404.514 | 6 / 0 | 4.6 | Yes |
| 43524 | 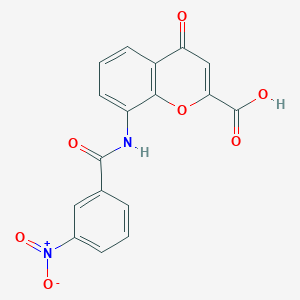 CHEMBL2392154 CHEMBL2392154 | C17H10N2O7 | 354.274 | 7 / 2 | 1.9 | Yes |
| 44140 | 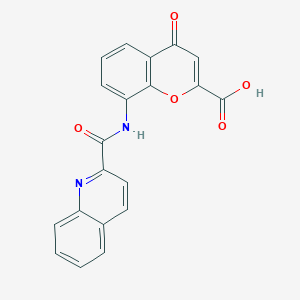 CHEMBL2392152 CHEMBL2392152 | C20H12N2O5 | 360.325 | 6 / 2 | 2.7 | Yes |
| 44268 | 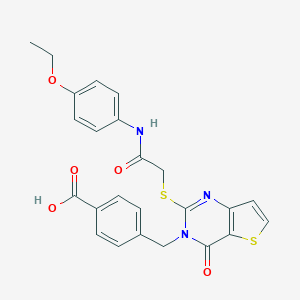 MLS-0435426.0001 MLS-0435426.0001 | C24H21N3O5S2 | 495.568 | 8 / 2 | 4.1 | Yes |
| 558711 | 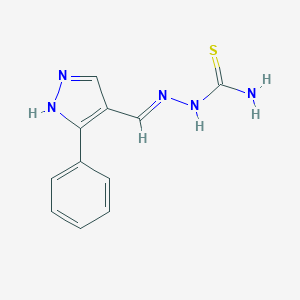 MLS-0437448.0001 MLS-0437448.0001 | C11H11N5S | 245.304 | 3 / 3 | 1.2 | Yes |
| 46615 |  MLS000575039 MLS000575039 | C24H22N2O5S2 | 482.569 | 7 / 3 | 5.1 | No |
| 46642 |  MLS000540783 MLS000540783 | C28H24N4O4S | 512.584 | 7 / 2 | 5.1 | No |
| 47056 |  CHEMBL2392175 CHEMBL2392175 | C19H14BrNO6 | 432.226 | 6 / 2 | 3.1 | Yes |
| 47349 |  CHEMBL2089226 CHEMBL2089226 | C10H8O5 | 208.169 | 5 / 3 | 1.8 | Yes |
| 47814 | 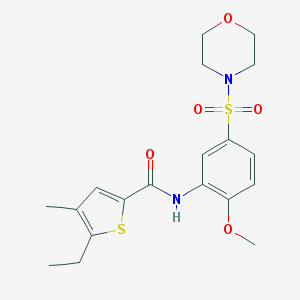 MLS-0418254.0001 MLS-0418254.0001 | C19H24N2O5S2 | 424.53 | 7 / 1 | 2.7 | Yes |
| 47844 | 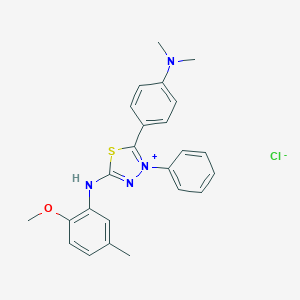 MLS000948484 MLS000948484 | C24H25ClN4OS | 453.001 | 6 / 1 | N/A | N/A |
| 48735 | 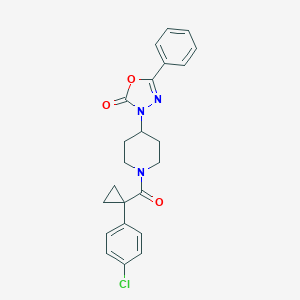 MLS001116134 MLS001116134 | C23H22ClN3O3 | 423.897 | 4 / 0 | 4.2 | Yes |
| 51438 | 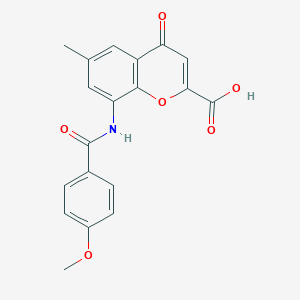 CHEMBL2425822 CHEMBL2425822 | C19H15NO6 | 353.33 | 6 / 2 | 2.5 | Yes |
| 53605 |  MLS-0004183.0001 MLS-0004183.0001 | C22H29ClN6O2 | 444.964 | 5 / 0 | 2.8 | Yes |
| 53707 |  264233-05-8 264233-05-8 | C17H19F2N5O2S | 395.429 | 7 / 2 | 2.9 | Yes |
| 53711 |  AC1MCK4S AC1MCK4S | C17H19F2N5O2S | 395.429 | 7 / 2 | 2.9 | Yes |
| 54744 |  MLS000834951 MLS000834951 | C14H13F2N5O2S | 353.348 | 7 / 2 | 2.0 | Yes |
| 54746 | 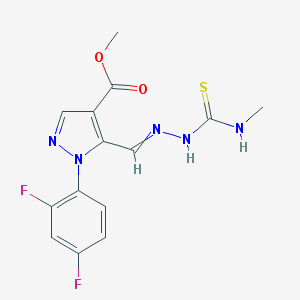 AC1MCK4M AC1MCK4M | C14H13F2N5O2S | 353.348 | 7 / 2 | 2.0 | Yes |
| 54808 | 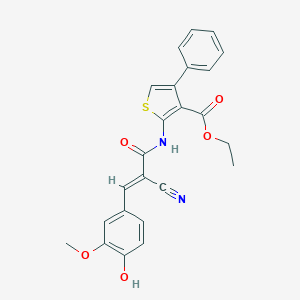 MLS000395028 MLS000395028 | C24H20N2O5S | 448.493 | 7 / 2 | 5.1 | No |
| 57629 | 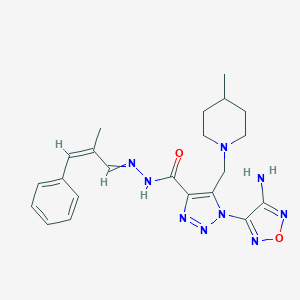 MLS000923861 MLS000923861 | C22H27N9O2 | 449.519 | 9 / 2 | 2.4 | Yes |
| 58548 | 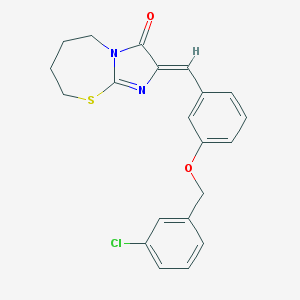 CHEMBL3221199 CHEMBL3221199 | C21H19ClN2O2S | 398.905 | 4 / 0 | 5.0 | Yes |
| 58735 | 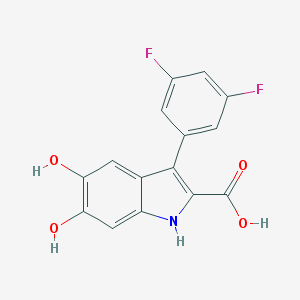 CHEMBL2089223 CHEMBL2089223 | C15H9F2NO4 | 305.237 | 6 / 4 | 3.0 | Yes |
| 59094 | 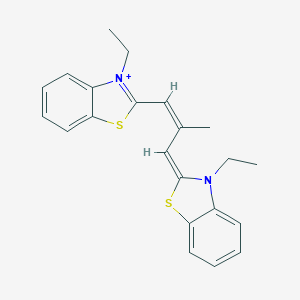 MCULE-8861643378 MCULE-8861643378 | C22H23N2S2+ | 379.56 | 3 / 0 | 6.9 | No |
| 59924 | 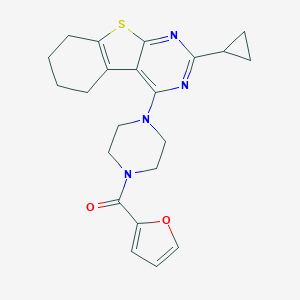 MLS000071807 MLS000071807 | C22H24N4O2S | 408.52 | 6 / 0 | 4.1 | Yes |
| 61621 | 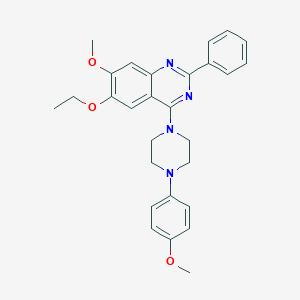 CHEMBL2431237 CHEMBL2431237 | C28H30N4O3 | 470.573 | 7 / 0 | 5.5 | No |
| 62877 | 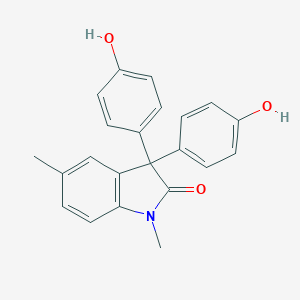 MLS000661102 MLS000661102 | C22H19NO3 | 345.398 | 3 / 2 | 3.8 | Yes |
| 63243 | 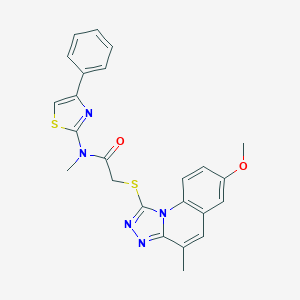 ASN 04482193 ASN 04482193 | C24H21N5O2S2 | 475.585 | 7 / 0 | 5.8 | No |
| 63620 | 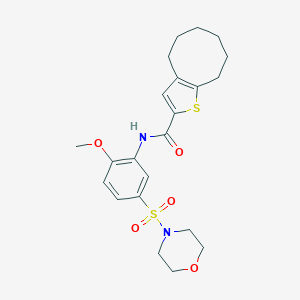 MLS-0435539.0001 MLS-0435539.0001 | C22H28N2O5S2 | 464.595 | 7 / 1 | 3.8 | Yes |
| 64701 | 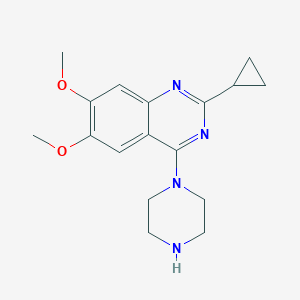 CHEMBL2431110 CHEMBL2431110 | C17H22N4O2 | 314.389 | 6 / 1 | 1.9 | Yes |
| 64865 |  dicumarol dicumarol | C19H12O6 | 336.299 | 6 / 2 | 2.6 | Yes |
| 64928 |  MLS-0433928.0001 MLS-0433928.0001 | C24H31N3O3S | 441.59 | 5 / 0 | 3.7 | Yes |
| 65215 |  MLS000862515 MLS000862515 | C28H25N5O4S | 527.599 | 8 / 2 | 4.4 | No |
| 65942 |  MLS001210236 MLS001210236 | C20H21IN2 | 416.306 | 1 / 0 | N/A | N/A |
| 66518 |  CHEMBL2392151 CHEMBL2392151 | C21H13NO5 | 359.337 | 5 / 2 | 3.4 | Yes |
| 66848 |  CHEMBL1914582 CHEMBL1914582 | C14H13N3O | 239.278 | 4 / 0 | 1.4 | Yes |
| 69501 |  MLS-0437152.0001 MLS-0437152.0001 | C21H20FN3O3 | 381.407 | 5 / 0 | 3.4 | Yes |
| 70338 |  MLS-0435540.0001 MLS-0435540.0001 | C27H36N4O3S | 496.67 | 6 / 0 | 3.9 | Yes |
| 70489 |  MLS-0437148.0001 MLS-0437148.0001 | C23H27N5O2 | 405.502 | 4 / 2 | 2.0 | Yes |
| 71471 | 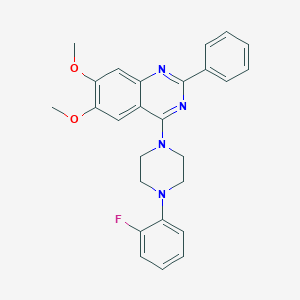 CHEMBL2431135 CHEMBL2431135 | C26H25FN4O2 | 444.51 | 7 / 0 | 5.3 | No |
| 71637 | 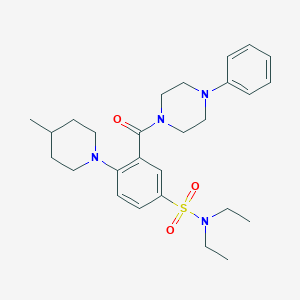 MLS-0435530.0001 MLS-0435530.0001 | C27H38N4O3S | 498.686 | 6 / 0 | 4.3 | Yes |
| 71839 | 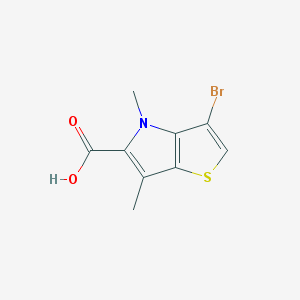 CHEMBL2037468 CHEMBL2037468 | C9H8BrNO2S | 274.132 | 3 / 1 | 2.7 | Yes |
| 73242 | 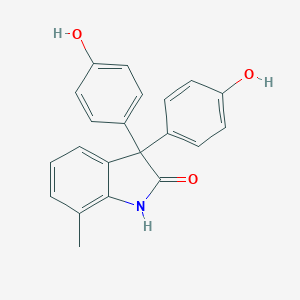 BHPI BHPI | C21H17NO3 | 331.371 | 3 / 3 | 3.6 | Yes |
| 74672 | 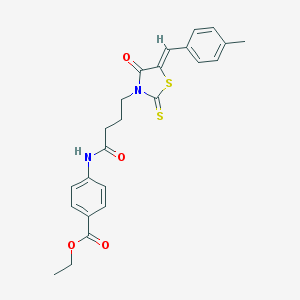 MLS-0435416.0001 MLS-0435416.0001 | C24H24N2O4S2 | 468.586 | 6 / 1 | 4.7 | Yes |
| 74878 | 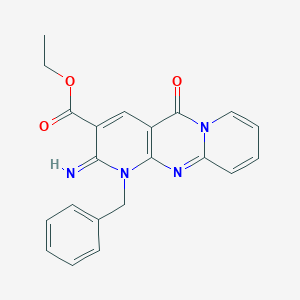 MLS000034919 MLS000034919 | C21H18N4O3 | 374.4 | 5 / 1 | 1.9 | Yes |
| 77049 | 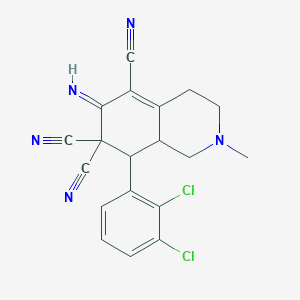 cid_655684 cid_655684 | C19H15Cl2N5 | 384.264 | 5 / 1 | 2.7 | Yes |
| 77347 | 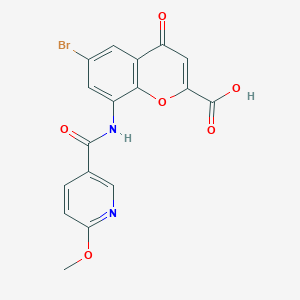 CHEMBL2392183 CHEMBL2392183 | C17H11BrN2O6 | 419.187 | 7 / 2 | 2.0 | Yes |
| 77571 | 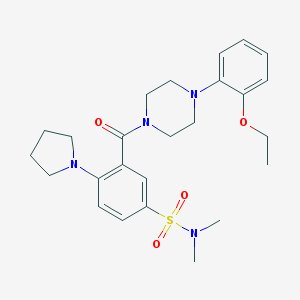 MLS-0421957.0001 MLS-0421957.0001 | C25H34N4O4S | 486.631 | 7 / 0 | 3.1 | Yes |
| 80581 | 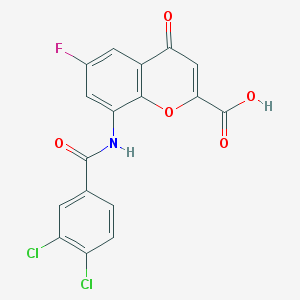 CHEMBL2392161 CHEMBL2392161 | C17H8Cl2FNO5 | 396.151 | 6 / 2 | 3.5 | Yes |
| 81351 | 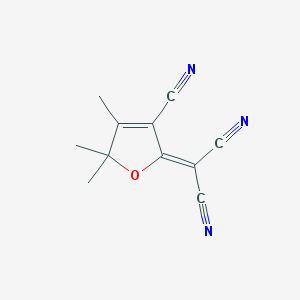 CHEMBL1914584 CHEMBL1914584 | C11H9N3O | 199.213 | 4 / 0 | 0.4 | Yes |
| 81838 | 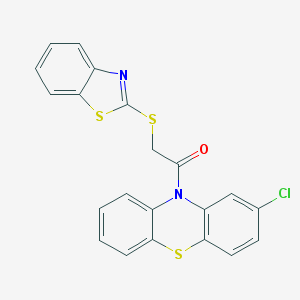 MLS000576788 MLS000576788 | C21H13ClN2OS3 | 440.978 | 5 / 0 | 6.6 | No |
| 82288 | 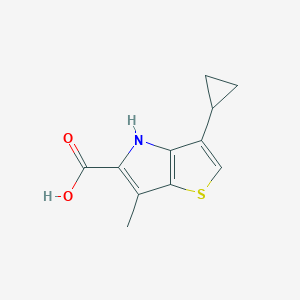 CHEMBL2037467 CHEMBL2037467 | C11H11NO2S | 221.274 | 3 / 2 | 3.0 | Yes |
| 83221 | 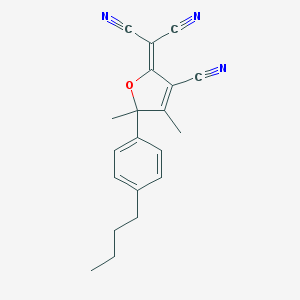 CHEMBL1914581 CHEMBL1914581 | C20H19N3O | 317.392 | 4 / 0 | 3.6 | Yes |
| 87246 | 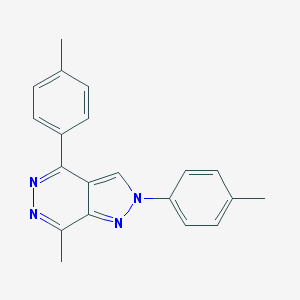 MLS-0437175.0001 MLS-0437175.0001 | C20H18N4 | 314.392 | 3 / 0 | 3.9 | Yes |
| 87490 | 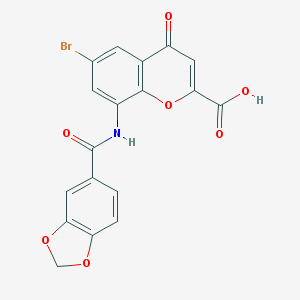 CHEMBL2392178 CHEMBL2392178 | C18H10BrNO7 | 432.182 | 7 / 2 | 2.6 | Yes |
| 560010 | 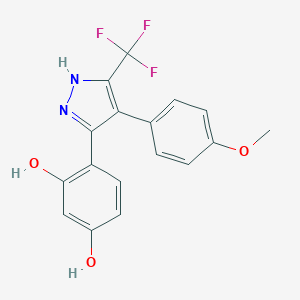 MLS000778974 MLS000778974 | C17H13F3N2O3 | 350.297 | 7 / 3 | 3.7 | Yes |
| 88584 | 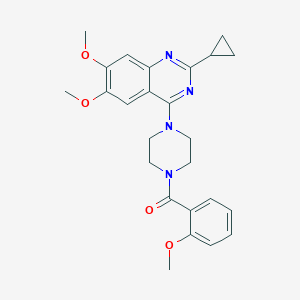 CHEMBL2431109 CHEMBL2431109 | C25H28N4O4 | 448.523 | 7 / 0 | 3.4 | Yes |
| 89238 | 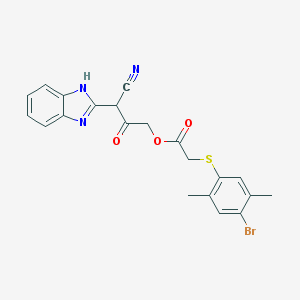 BDBM54304 BDBM54304 | C21H18BrN3O3S | 472.357 | 6 / 1 | 4.7 | Yes |
| 89756 | 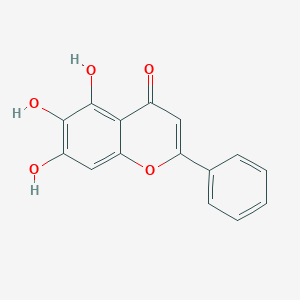 baicalein baicalein | C15H10O5 | 270.24 | 5 / 3 | 1.7 | Yes |
| 90292 | 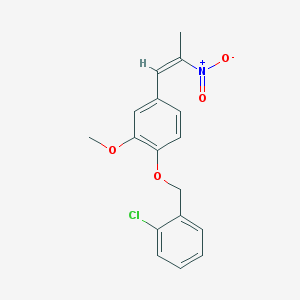 SMR000203542 SMR000203542 | C17H16ClNO4 | 333.768 | 4 / 0 | 4.6 | Yes |
| 92116 | 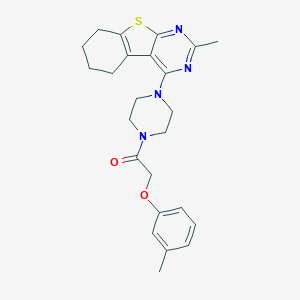 MLS-0437133.0001 MLS-0437133.0001 | C24H28N4O2S | 436.574 | 6 / 0 | 5.0 | Yes |
| 92957 | 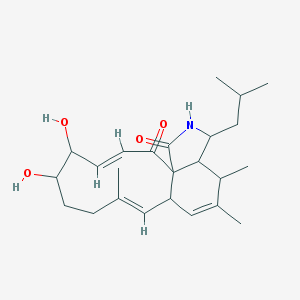 MLS000876956 MLS000876956 | C24H35NO4 | 401.547 | 4 / 3 | 2.5 | Yes |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218